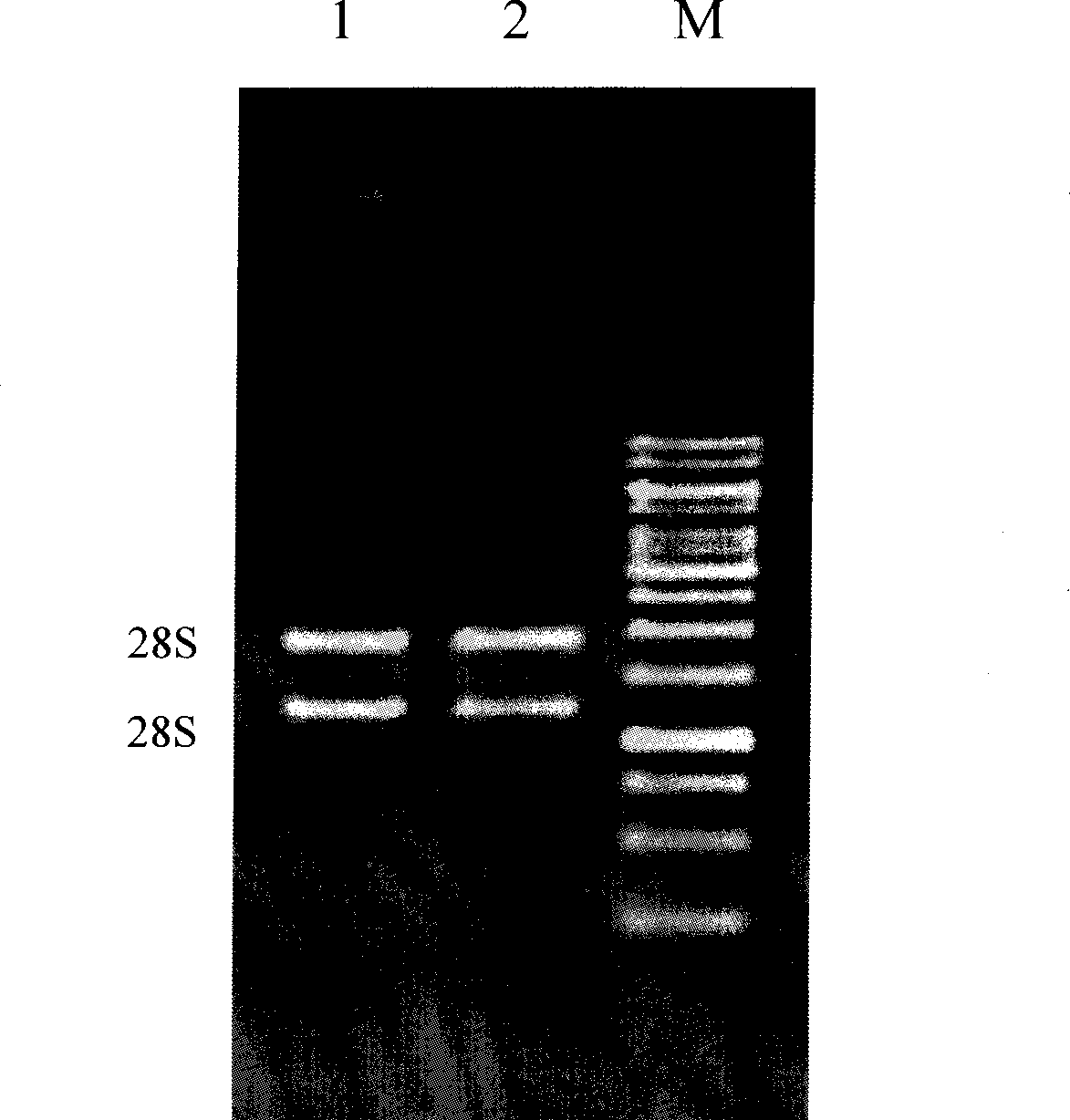
RNA extraction method by using silicon film pole to adsorb RNA
An extraction method and column adsorption technology, which is applied in the field of RNA extraction using silicon membrane column to absorb RNA, can solve the problems of DNA pollution, shorten the extraction time, etc., and achieve the effect of simple formula, shortened extraction time and high purity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Preparation of denaturing solution:
[0058] ①Take an 80ml bottle, weigh 3.65g of solid guanidine isothiocyanate, and put it into the bottle;
[0059] ②Weigh 1.54g sodium citrate and put it into the bottle;
[0060] ③ Dilute to 10ml with double distilled water;
[0061] ④Heat in a water bath at 60°C until completely dissolved, then adjust the pH to 4.0 with citric acid;
[0062] ⑤Store at room temperature for later use.
[0063] Steps to extract RNA:
[0064]①Add 1000 μl denaturing solution and 240 μl chloroform to the prepared Pseudomonas strain M18, vortex and mix well, centrifuge at 12,000 rpm at 4°C for 1 min, and the bacterial solution is divided into three layers in the tube;
[0065] ② Transfer 500 μl of the upper liquid to a silicon membrane column, add 250 μl of isopropanol, and centrifuge at 12,000 rpm for 1 min;
[0066] ③ Add 500 μl of 75% ethanol washing solution to the silicon membrane column, and centrifuge at 12,000 rpm for 1 min;
[0067] ④Repeat ...
Embodiment 2
[0073] Preparation of denaturing solution:
[0074] The steps are the same as in Example 1, wherein: 3.65g sodium lauryl sarcosine, 2.95g sodium acetate, dilute to 10ml with double distilled water, heat in a 60°C water bath until completely dissolved, then adjust the pH to 4.0 with pure acetic acid. Set aside at room temperature.
[0075] Steps to extract RNA:
[0076] ①Add 1000 μl denaturing solution and 240 μl chloroform to the prepared yeast cells, vortex and mix well, centrifuge at 12,000 rpm at 4°C for 1 min, and the bacterial solution is divided into three layers in the tube;
[0077] ② Transfer 500 μl of the upper layer liquid to a silicon membrane column, and centrifuge at 12,000 rpm for 1 min;
[0078] ③ Add 500 μl of 75% ethanol washing solution to the silicon membrane column, and centrifuge at 12,000 rpm for 1 min;
[0079] ④Repeat the previous step;
[0080] ⑤Centrifuge at 12,000rpm for 1min to ensure that there is no liquid in the silicon membrane column;
[...
Embodiment 3
[0085] Preparation of denaturing solution:
[0086] The procedure is the same as in Example 1, wherein: 2ml of saturated phenol, 3.05g of sodium lactate, and distilled water to 10ml. Heat in a water bath at 60°C until completely dissolved, then adjust the pH to 4.0 with lactic acid. Set aside at room temperature.
[0087] Steps to extract RNA:
[0088] ① Add 1000 μl denaturing solution and 240 μl chloroform to the prepared fresh tea sample, vortex and mix well, centrifuge at 12,000 rpm at 4°C for 1 min, and the bacterial solution is divided into three layers in the tube;
[0089] ② Transfer 500 μl of the upper liquid to a silicon membrane column, add 250 μl of isopropanol, and centrifuge at 12,000 rpm for 1 min;
[0090] ③ Add 500 μl of 75% ethanol washing solution to the silicon membrane column, and centrifuge at 12,000 rpm for 1 min;
[0091] ④Repeat the previous step;
[0092] ⑤Centrifuge at 12,000rpm for 1min to ensure that there is no liquid in the silicon membrane c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com