Method for constructing and producing restriction endonuclease Ecop15I
A restriction endonuclease and enzyme linking technology, applied in the field of bioengineering, can solve the problems of poor expression effect and affect the biological activity of protein complexes, and achieve the effect of good application value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Embodiment 1, material and reagent
[0048] 1. Materials:
[0049] Plasmids and cell lines pUC19 and pUC57 are products of Biotech (Nantong) Co., Ltd. (Nantong Biotech). Root Biotechnology Ltd., based on Molecular Cloning CaCl 2 Self-made competent state, guarantee titer above 106 (CFU / ug pUC19).
[0050] 2. Reagents:
[0051] (1) Tool enzymes: Taq DNA polymerase, Pfu DNA polymerase (Nantong Biolabs); restriction enzymes, SmaI, BamHI, etc. (New England Biolabs); T4 DNA ligase (New England Biolabs);
[0052] (2) Biochemical reagents: Yeast, Typtone, Tris base, glycine, SDS, agarose, etc. (Shanghai Sangong);
[0053] (3) DNA purification kit: common type plasmid recovery kit, agarose gel DNA recovery kit (Nantong Biotech);
[0054] (4) Chromatography packing: Ni sepharose affinity packing; DEAE sepharose anion exchange packing; CM sepharose anion exchange packing (Beijing Webster Bohui).
Embodiment 2
[0055] Embodiment 2, experimental method
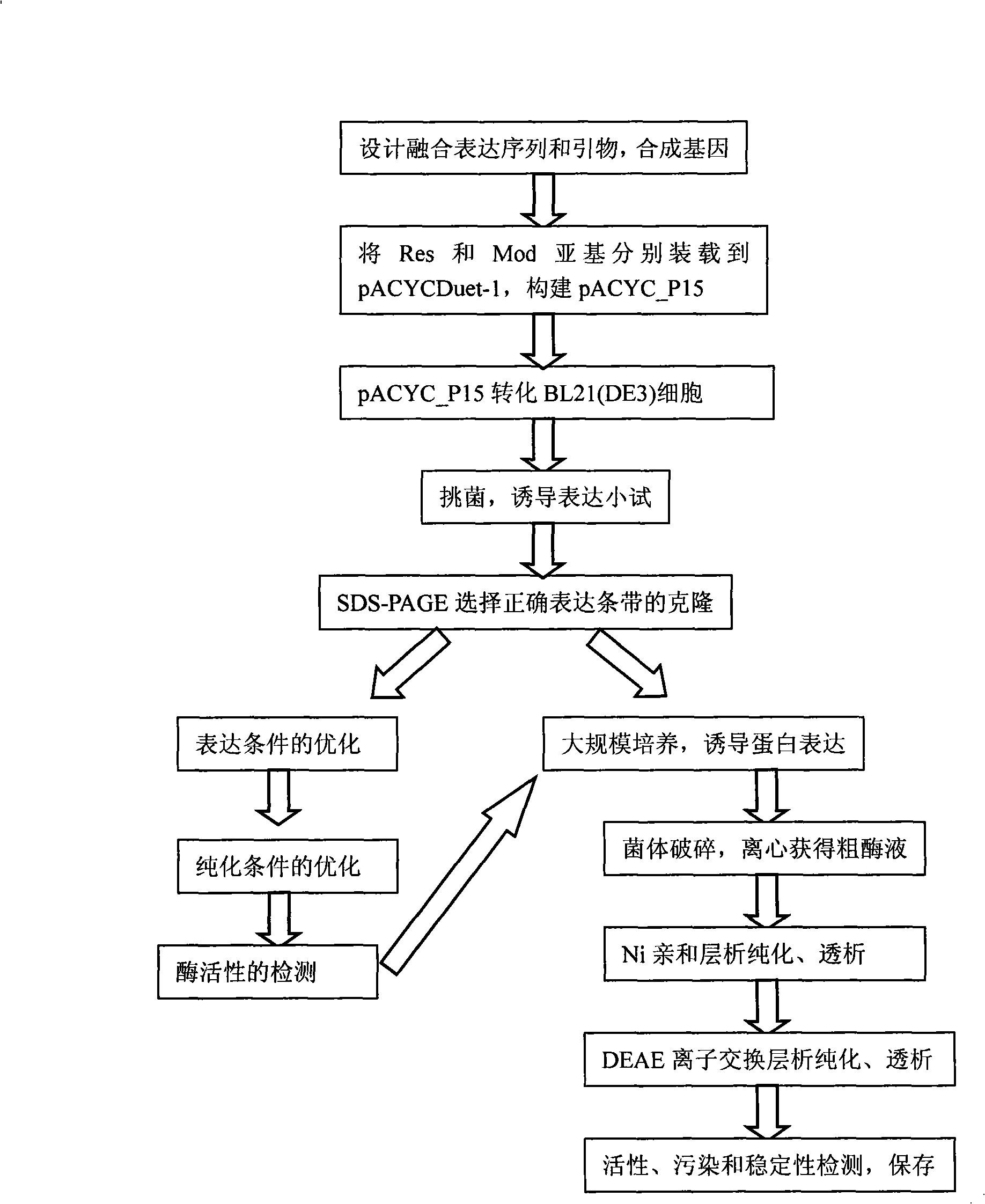
[0056] 1. Artificial synthesis of genes
[0057] The Ecop15I gene sequence (Genbank No. AJ634453) was obtained from the NCBI website, and the fusion expression sequence was designed. Considering the convenience of downstream enzyme protein purification requirements, 6×His Tag was fused to the C-terminus of the Res subunit and the N-terminus of the Mod subunit of Ecop15I, respectively.
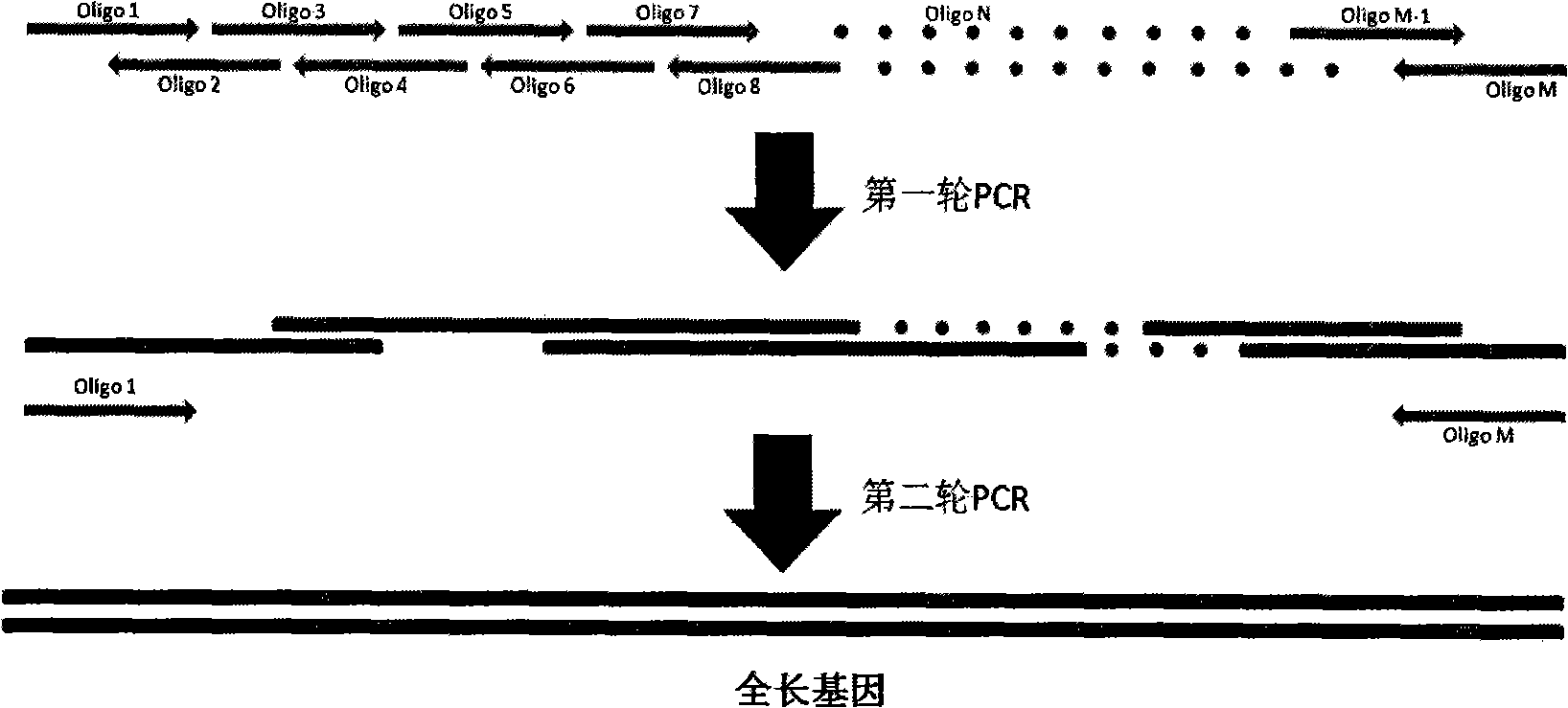
[0058] Dozens of pairs of primers were designed according to the principle of overlapping oligonucleotides. KpnI, XhoI, BamHI, and SalI restriction sites were added to the head and tail of the Res and Mod fragments of the primers at both ends, and a G was added after the BamHI restriction site. bases to ensure that the enzyme sequence is in the correct reading frame during translation. For the nearly 3kb Res gene fragment, the Res fragment is divided into two segments through the only BglII restriction site in the middle, and then digested with KpnI ...
Embodiment 3
[0069] Embodiment 3, experimental result
[0070] (1) Construction results of pACYC_P15 co-expression plasmid
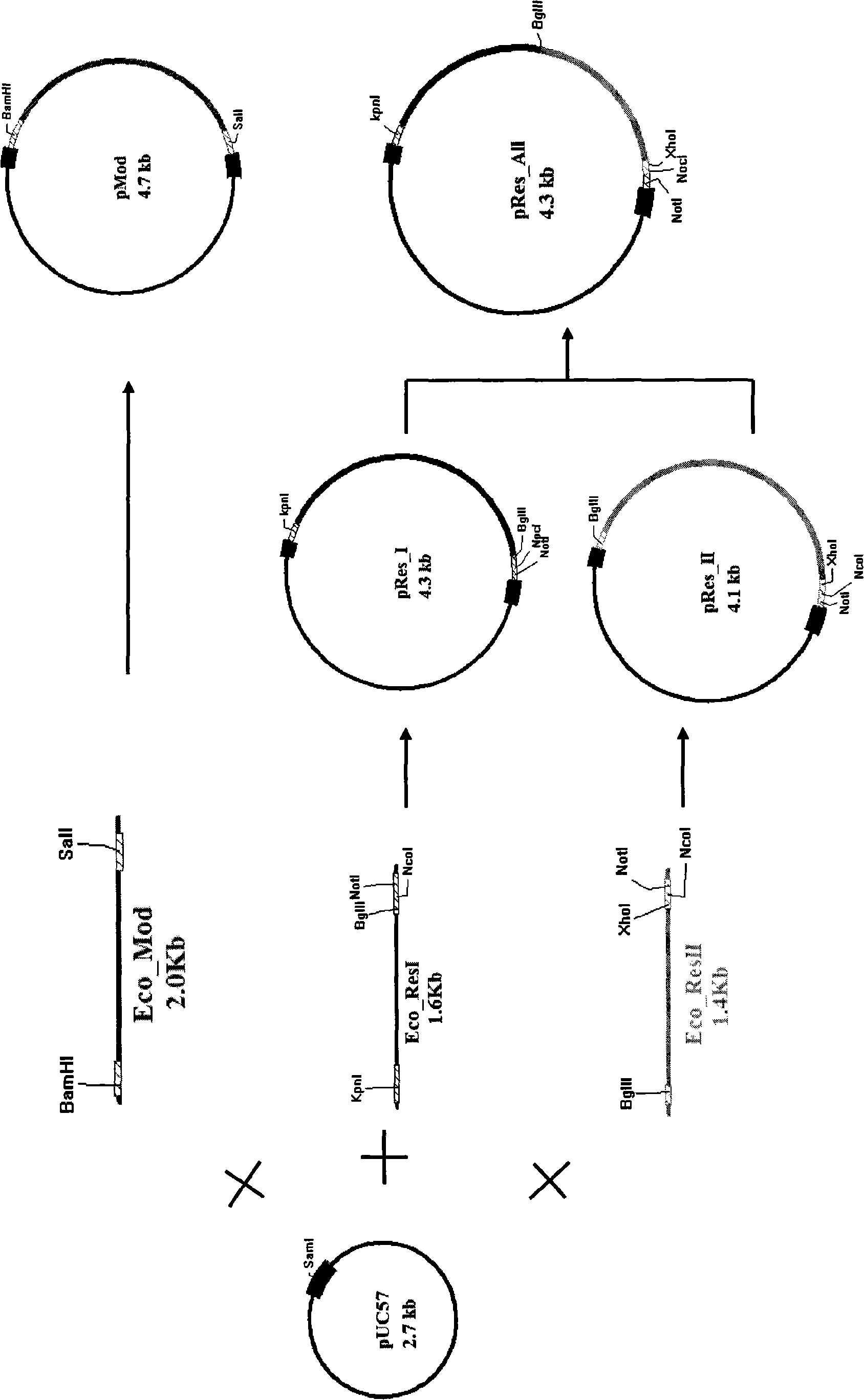
[0071] The Res subunit and the Mod subunit of Ecop15I synthesized 3 genes by artificial PCR, respectively Eco_Mod, Eco_ResI and Eco_ResII (see attached Figure 3 Show). The three genes were respectively blunt-ended cloned and loaded into pUC57 to obtain three plasmids, pMod, pRes_I and pRes_II; pRes_I and pRes_II were digested with BglII and NotI, and ligated with T4 ligase to obtain the complete Res fragment plasmid.
[0072] The pMod plasmid was digested with the pACYCDuet-1 plasmid BamHI and SalI and then enzyme-ligated to obtain the pACYC_Mod plasmid containing the Mod subunit, and then combined with the pRes_All plasmid KpnI and XhoI to obtain the two complete Ecop15I subunits pACYC_P15 plasmid (see attached Figure 4 Show).
[0073] (2) Expression and purification of Ecop15I
[0074] Using the induction time gradient, IPTG induction concentration gradient,...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com