High-specific-activity xylanase XYN11F63 and genes and application thereof
A technology of XYN11F63 and xyn11f63, applied in the field of genetic engineering, can solve the problems of limited application and high production cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
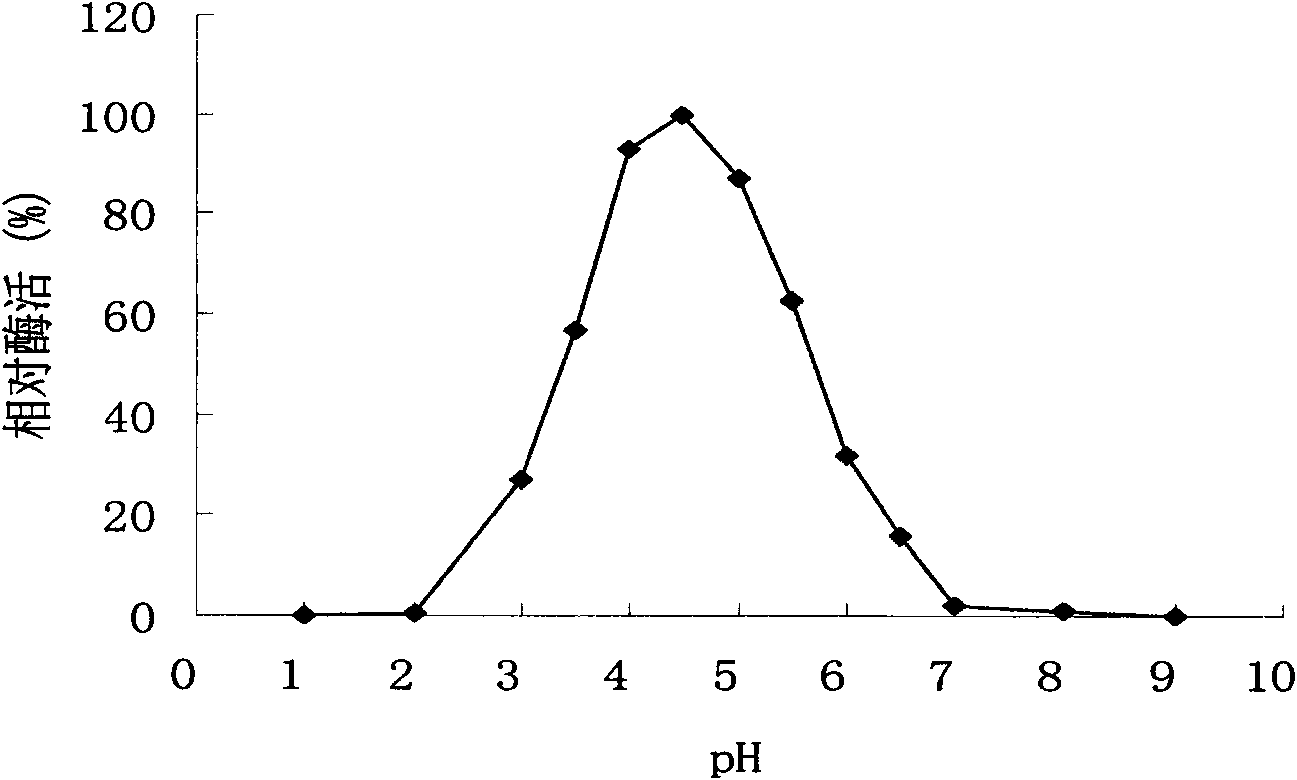
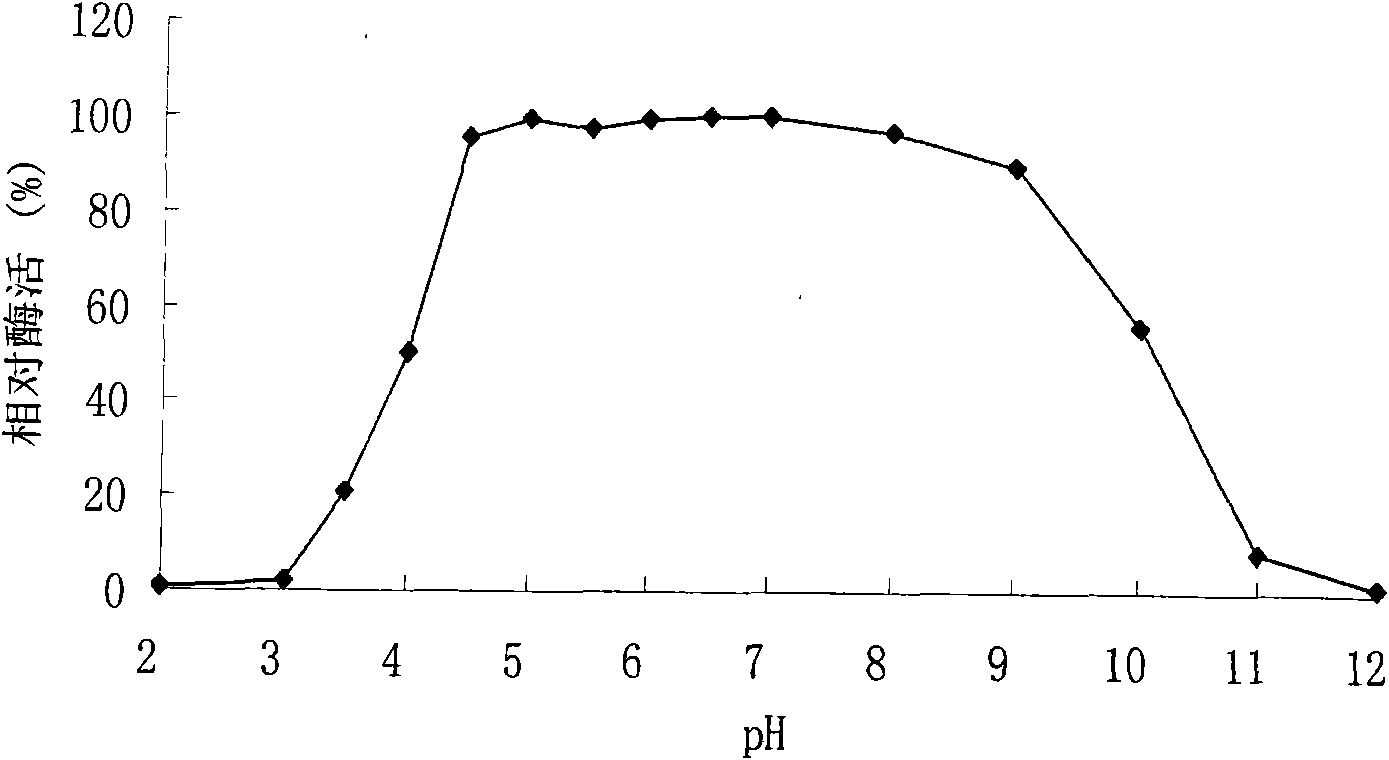
[0055] Example 1 Enzyme production characteristics of Penicillium sp. F63 CGMCC1669
[0056] The Penicillium sp.F63 CGMCC1669 was cultured in potato juice medium and then spread on the enzyme production medium ((NH 4 ) 2 SO 4 5g / L, KH 2 PO 4 1g / L, MgSO 4 ·7H 2 O 0.5g / L, FeSO 4 ·7H 2 O0.01g / L, CaCl 2 0.2g / L, 1% xylan, 1.5% agarose, pH 5.0) plate, cultured at 30°C for 5-6 days, transparent circles can be seen on the plate with enzyme production medium. Prove that it has xylanase activity.
Embodiment 2
[0057] Example 2 Cloning of Penicillium sp.F63 (CGMCC1669) xylanase encoding gene xyn11F63
[0058] Extract the genomic DNA of Penicillium sp.F63 (CGMCC1669):
[0059] Filter the mycelium cultured for 3 days with sterile filter paper and put it in a mortar, add 2 mL of extract, grind for 5 min, then place the grind in a 50 mL centrifuge tube, lyse in a water bath at 65°C for 20 min, and mix every 10 min. Homogenize once and centrifuge at 10,000 rpm at 4°C for 5 min. The supernatant was extracted in phenol / chloroform to remove impurities, and then the supernatant was added to an equal volume of isopropanol. After standing at room temperature for 5 min, centrifuged at 10,000 rpm at 4°C for 10 min. The supernatant was discarded, the precipitate was washed twice with 70% ethanol, dried in vacuum, dissolved in an appropriate amount of TE, and placed at -20°C for use.
[0060] Degenerate primers P1, P2 were designed and synthesized according to the conservative (CYLG(A)VYGW and TFNQYWS) ...
Embodiment 3
[0069] Example 3 RT-PCR analysis of xylanase gene
[0070] Extract the total RNA of Penicillium sp.F63 (CGMCC166), use reverse transcriptase to obtain a strand of cDNA, and then design the appropriate primers (Xy110A F: 5'-ATGGTCTCTTTTTCAAACCTCTTTATGGCTGCCTG-3', Xy110A R: 5'-TTAGGAAACAGTGATGGACGAAGAGCCACT-3' ) Amplify the single-stranded cDNA to obtain the cDNA sequence of xylanase, and the amplified product is recovered and sent to Sanbo Biotechnology Co., Ltd. for sequencing.
[0071] By comparing the genome sequence of xylanase with the cDNA sequence, it was found that the gene has an intron. The cDNA is 654 bp in length, encoding 217 amino acids and a stop codon, and the N-terminal 19 amino acids are its predicted signal peptide sequence. The detected partial nucleotide sequence of the mature protein of the gene xyn11F63 was compared with the xylanase gene sequence on GeneBank. The highest identity was 86%, and the highest identity of the amino acid sequence was 85%, which prov...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com