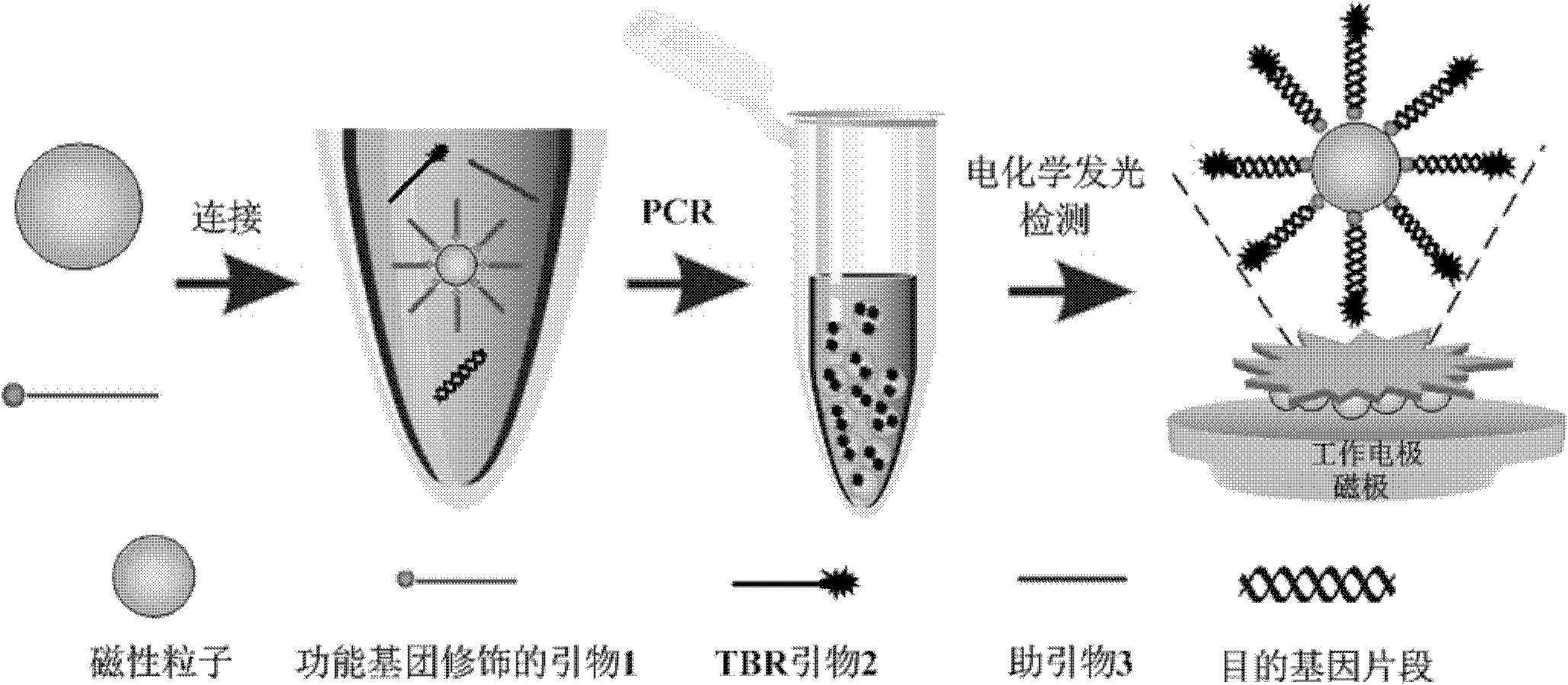
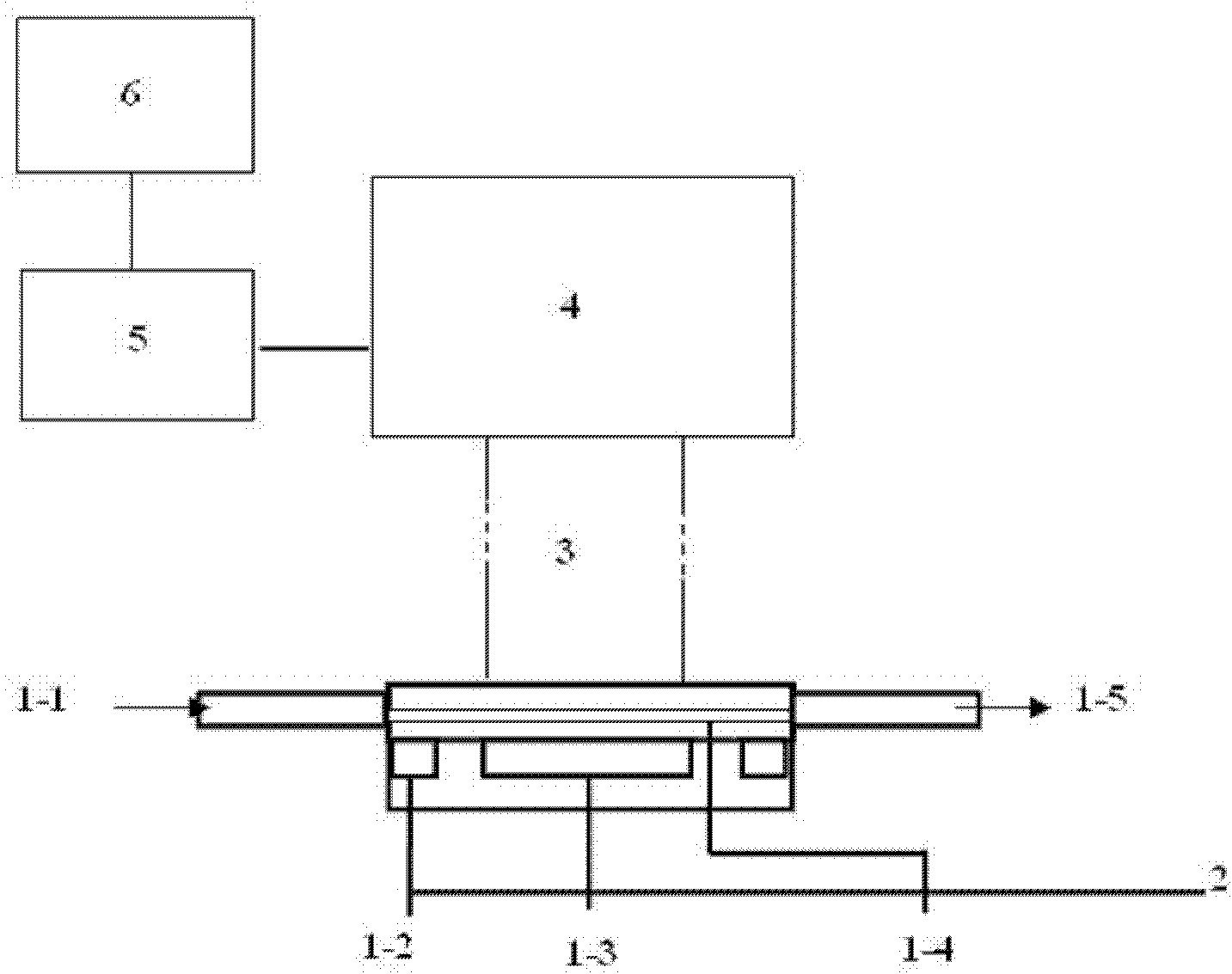
Method for detecting food pathogen by electrochemical luminescence gene sensor on basis of magnetic in-situ amplification
A technology of luminescent genes and pathogenic bacteria, applied in the field of electrochemiluminescence gene detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] (1) According to conventional methods, use TIANamp Bacteria DNA Kit to extract DNA from milk samples;
[0060] (2) Take 1 mg of magnetic particles with a particle size of 100 nm and surface modified with carboxyl groups, and wash them with 200 μL of 0.01M sodium hydroxide solution at room temperature for 2 times, and then rinse with deionized water once.
[0061] (3) To the washed magnetic particles, add a highly specific 5'end modified primer 1 with an amino group (see Table 1 for the sequence) 2nmol and 100μL pH5.0 ligation reaction solution (0.1M 2-? Incubate for 5 hours at room temperature with phytoethanesulfonic acid (MES), 0.25% Tween-20, 10mM carbodiimide hydrochloride (EDC), and shake the mixer slightly.
[0062] (4) After the reaction is completed, use a magnetic separator to separate the prepared magnetic primers, and wash them twice with a washing buffer (1 times standard 10 mM, pH 7.4 PBS, 0.25% Tween-20). Finally, the synthesized magnetic primers were placed in ...
Embodiment 2
[0072] Example 2 Detection sensitivity test of Listeria monocytogenes
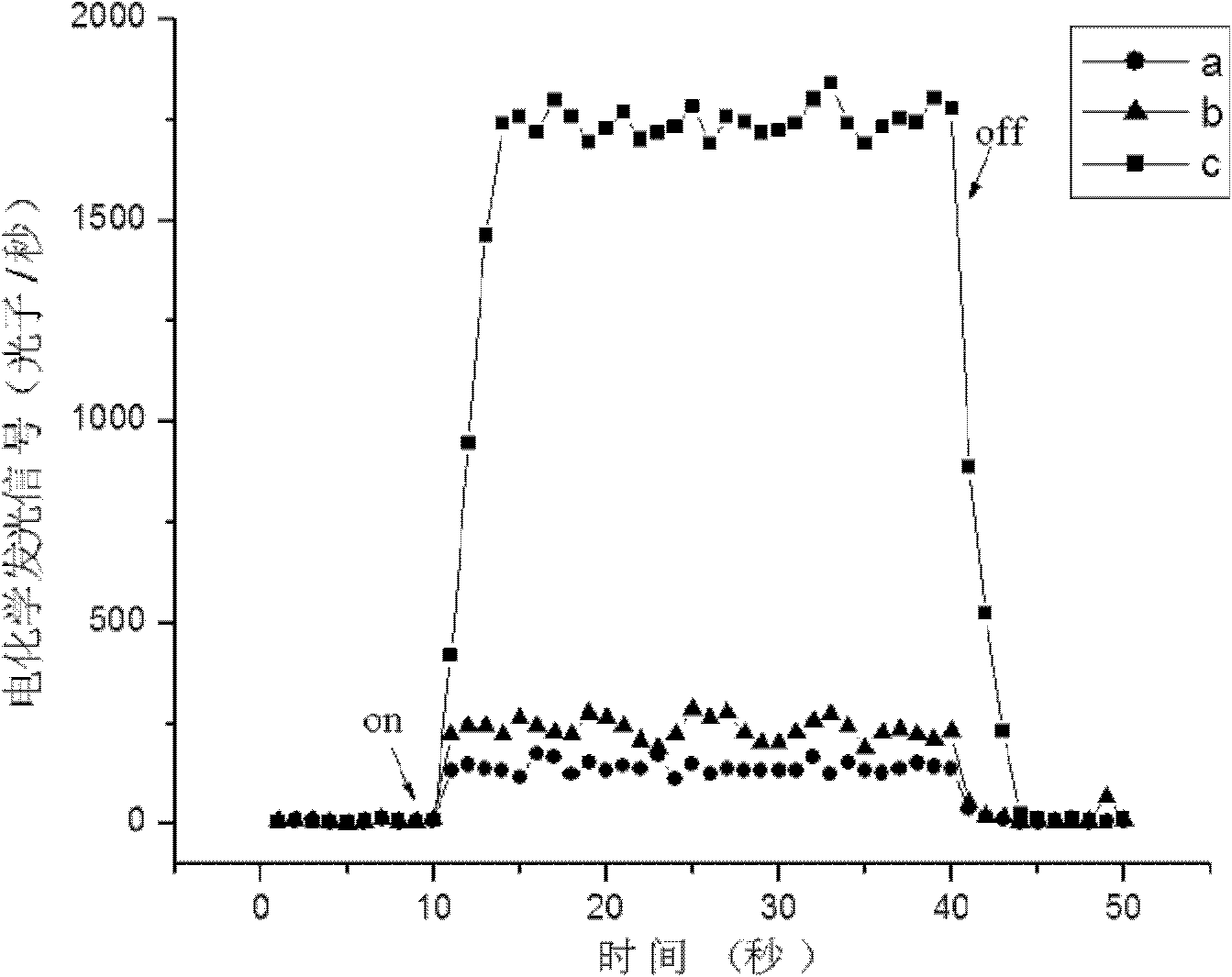
[0073] Take 10 respectively 5 pg, 10 4 pg, 10 3 pg, 10 2 pg, 50pg, 10pg Listeria monocytogenes (CMCC54007) genome, the electrochemical signal value measured according to the method described in Example 1 is as follows Figure 4 Shown by Figure 4 It can be seen that when the concentration of the target genome fragment of Listeria monocytogenes is equal to 0.5pg / ul, the electrochemiluminescence signal is still greater than the threshold 299cps (the luminescence average of the three groups of negative samples is 230cps, and the 3-fold standard deviation between samples is 66cps), so it can It is judged that the detection target gene of the present invention has very good sensitivity.
Embodiment 3
[0074] Example 3 Electrochemiluminescence verification experiment for the specificity of Listeria monocytogenes detection
[0075] Take 100ng Listeria monocytogenes (CMCC54007) genome, 100ng Escherichia coli (E.coli O157:H7, GW1.2020) genome, and 100ng Salmonella enterica (CMCC50040) genome respectively, according to the method described in Example 1. The measured electrochemical signal value is as Figure 5 Shown by Figure 5 It can be seen that when the genomes of Escherichia coli, Salmonella and Listeria monocytogenes at the same mass concentration are respectively present in the sample, only when the target genome is present can a higher electrochemiluminescence signal be obtained, which can be judged by the method of the present invention. Listeria monocytogenes has very good specificity.
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com