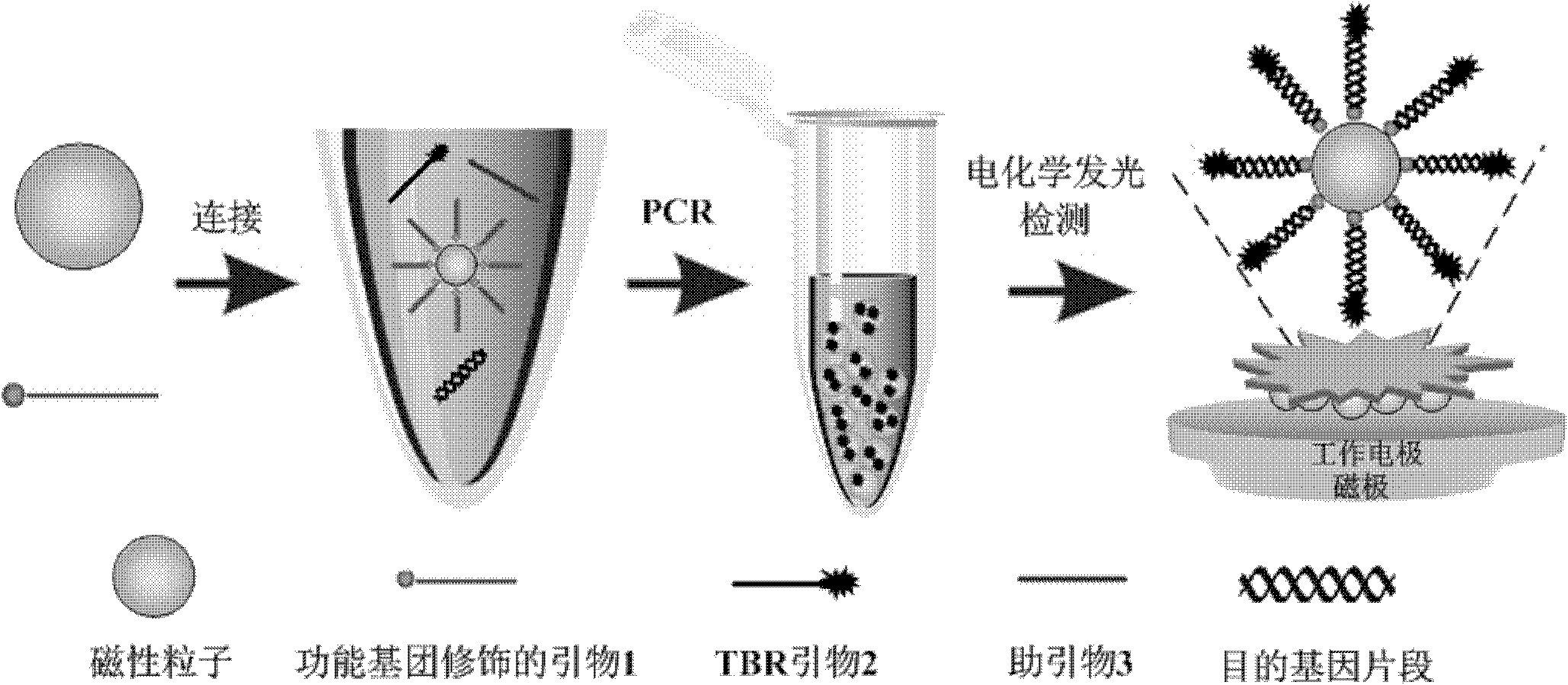
Method for detecting food pathogen by electrochemical luminescence gene sensor on basis of magnetic in-situ amplification
A technology of luminescent genes and pathogenic bacteria, applied in the field of electrochemiluminescence gene detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] (1) According to the conventional method, use the TIANamp Bacteria DNA Kit kit to extract the DNA in the milk sample;
[0060] (2) Take 1 mg of magnetic particles with a particle size of 100 nm and surface-modified carboxyl groups, wash them twice with 200 μL of 0.01 M sodium hydroxide solution at room temperature, and then wash them once with deionized water.
[0061] (3) Add 2 nmol of a designed highly specific 5'-end amino-modified primer 1 (see Table 1 for the sequence) and 100 μL of ligation reaction solution (0.1M 2-? Methyrethanesulfonic acid (MES), 0.25% Tween-20, 10 mM carbodiimide hydrochloride (EDC)) were co-incubated for 5 hours at room temperature with a shaker gently shaking.
[0062] (4) After the reaction was completed, the prepared magnetic primers were separated using a magnetic separation pen, and washed twice with washing buffer (1 times standard 10 mM, PBS pH 7.4, 0.25% Tween-20). Finally, the synthesized magnetic primers were placed in 100 μL stor...
Embodiment 2
[0072] Example two Listeria monocytogenes detection sensitivity experiment
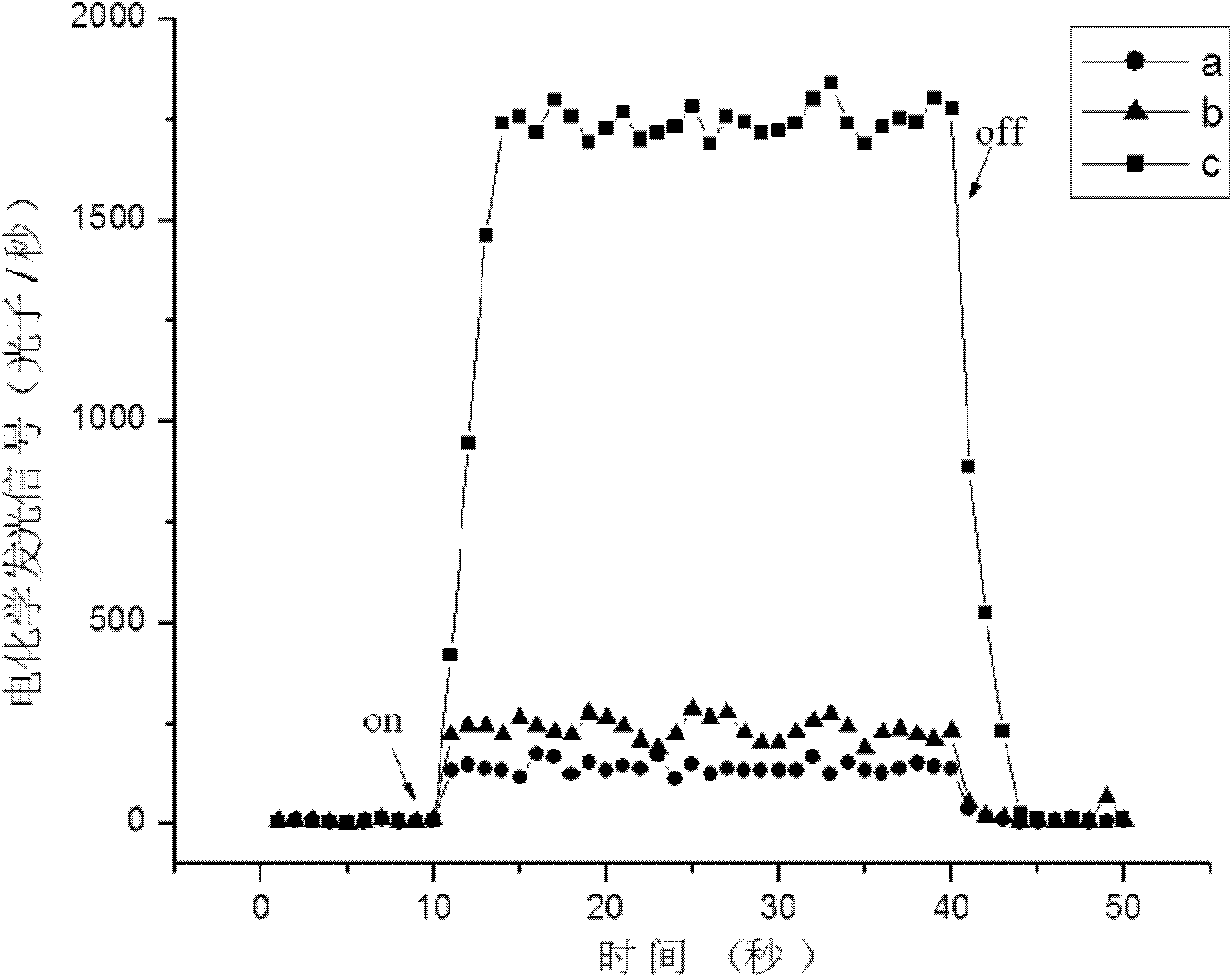
[0073] Take 10 respectively 5 pg, 10 4 pg, 10 3 pg, 10 2 pg, 50pg, 10pg Listeria monocytogenes (Listeria monocytogenes, CMCC54007) genome, according to the method described in Example 1, the electrochemical signal value is measured as Figure 4 shown by Figure 4 It can be seen that when the concentration of the genome fragment of Listeria monocytogenes is equal to 0.5pg / ul, the electrochemiluminescence signal is still greater than the threshold value of 299cps (the average value of the luminescence of the three negative samples is 230cps, and the three-fold standard deviation between samples is 66cps), so it can be It is judged that the detection target gene of the present invention has very good sensitivity.
Embodiment 3
[0074] Example 3 Electrochemiluminescence verification experiment of detection specificity of Listeria monocytogenes
[0075]Get respectively 100ng Listeria monocytogenes (Listeria monocytogenes, CMCC54007) genome, 100ng Escherichia coli (E.coli O157:H7, GW1.2020) genome, 100ng Salmonella (Salmonella enterica, CMCC50040) genome, according to the method described in Example 1 The measured electrochemical signal values are as Figure 5 shown by Figure 5 It can be seen that when there are Escherichia coli, Salmonella and Listeria monocytogenes genomes of the same mass concentration in the sample, only when the target genome exists, can a higher electrochemiluminescence signal be obtained, thus it can be judged that the method of the present invention can detect Listeria monocytogenes has very good specificity.
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com