Fluorescent PCR (polymerase chain reaction) detection kit for vibrio cholerae and systematic identification method thereof
A detection kit and Vibrio cholerae technology, applied in the direction of microorganism-based methods, biochemical equipment and methods, microorganism measurement/inspection, etc., can solve the problems of enhanced toxicity, unstable results, long cycle, etc., and achieve saving detection The effect of reducing cost, shortening detection time, and improving detection rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
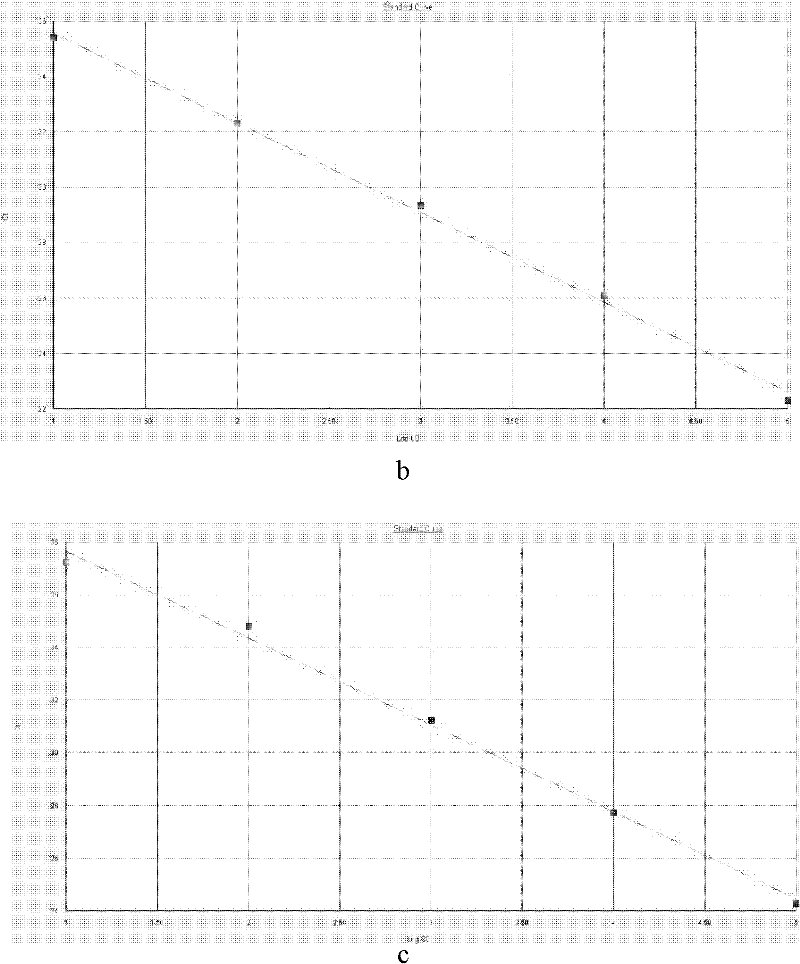
Image
Examples
Embodiment 1
[0058] Embodiment 1: the acquisition of specific primers, fluorescent probes and standards
[0059] 1. Materials:
[0060] Bacterial genomic DNA extraction reagents were purchased from Treasure Bioengineering (Dalian) Co., Ltd.; pGEM-T-Easy cloning system and Taq DNA polymerase were purchased from Promega Company of the United States; sequencing reagents and 377 sequencer were all products of ABI Company of the United States. Spectrophotometer (Eppendorf Company), Bio-Rad icycler PCR instrument (Bio-Rad Company) and ABI7500FAST type quantitative PCR instrument (ABI Company)
[0061] 2. Primer and probe design and synthesis:
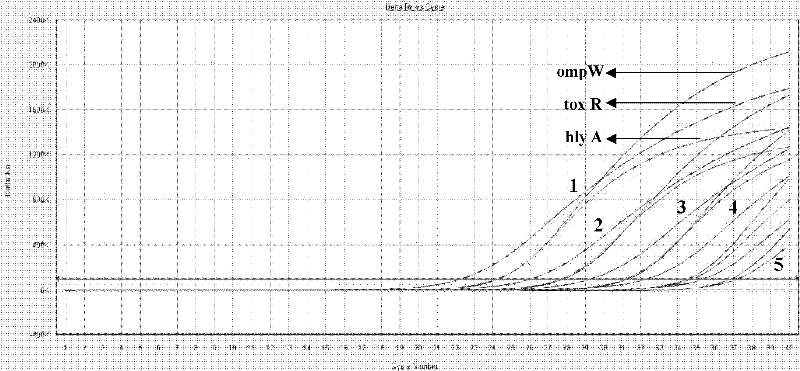
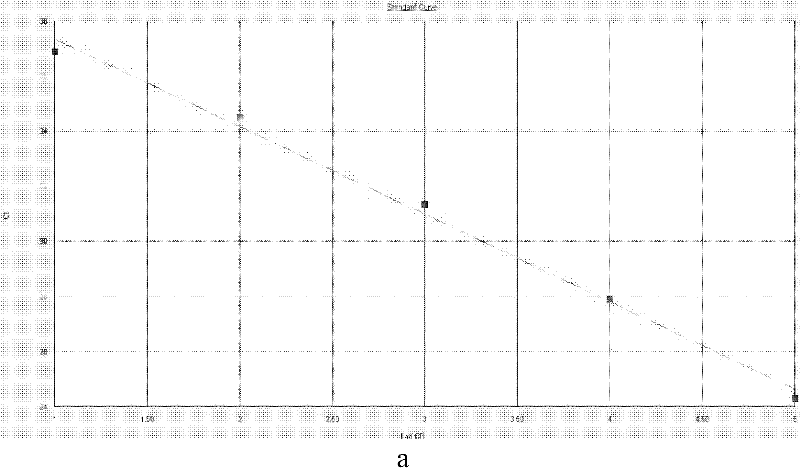
[0062] Using Vibrio cholerae outer membrane protein gene (ompW) (registration number DQ776044), toxin expression regulatory protein gene (toxR) (registration number HQ452873) and hemolysin gene (hlyA) (registration number GU230682) as templates, use Primer Express TM (V2.0, American ABI Company) software analyzes TaqMan primer and probe site, selects t...
Embodiment 2
[0086] Example 2: Systematic identification of Vibrio cholerae and its related toxin gene detection
[0087] 1. Identification of isolates from environmental samples:
[0088] Inoculate 30 bacterial strains to be identified on Columbia agar medium, culture in a 37°C incubator for 18-24 hours, pick a colorless, transparent, round, smooth and moist colony and puncture it on a three-sugar slant, culture it at 37°C for 18-24 hours Observe the results, and at the same time, the above-mentioned colonies are tested for oxidase, sticky silk and unsalted peptone water.
[0089] Santang slant: Colorless, transparent, round, smooth and moist colonies are punctured on the three sugar slant, cultured at 37°C for 18-24 hours, and the results are observed. The red and yellow on the slant are positive;
[0090] Sticky silk test: drop a large drop (about 15 μl) of the reagent on a glass slide or a plate, then take 1 loop of fresh agar culture of the tested bacteria, put it next to the reagent...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com