Production of recombinant mixed isoamylases, alpha amylases and glucoamylases
A technology of glucoamylase and alpha amylase, applied in the biological field, can solve problems such as strains and mixed enzyme products that have not yet mixed expression enzymes, and achieve the effect of saving raw materials
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
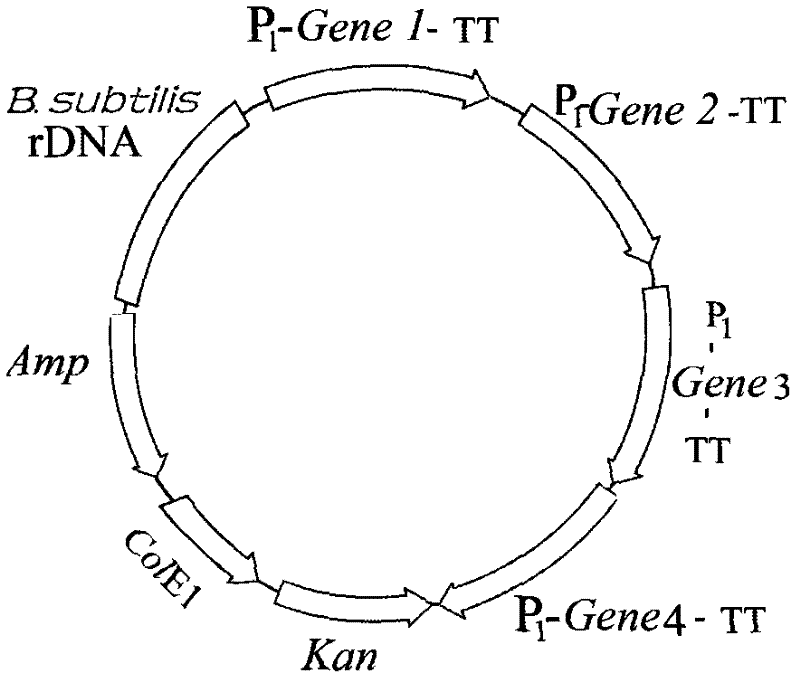
[0020] Example 1: Construction of Bacillus subtilis engineering bacteria to produce recombinant mixed isoamylase, α-amylase and glucoamylase
[0021] 1.1 Construction of Bacillus subtilis expression vector
[0022] 1.1.1 Construction of cloning vector
[0023] DNA synthesis contains ampicillin (AMP) gene sequence, polyclonal adapter and Escherichia coli replication origin, forming base complementarity at both ends of the sequence. It is circularized by the action of DNA ligase to form a DNA cloning vector. The cloning vector was named pBPA.
[0024] 1.1.2 Acquiring genes
[0025] ①PCR amplification of isoamylase gene
[0026] Genomic DNA of Pseudomonas amyloliquefaciens was extracted, and primers (5'CGGATATC GCCATCAACAGCATGAGC3'; 5'CGGATATCGGCTACTTGGAGATCAACAG3') were used for PCR amplification. The PCR product was sequence determined and analyzed with BLAST software provided by NCBI, which proved to be the sequence of the isoamylase gene.
[0027] ②PCR amplification of α...
Embodiment 2
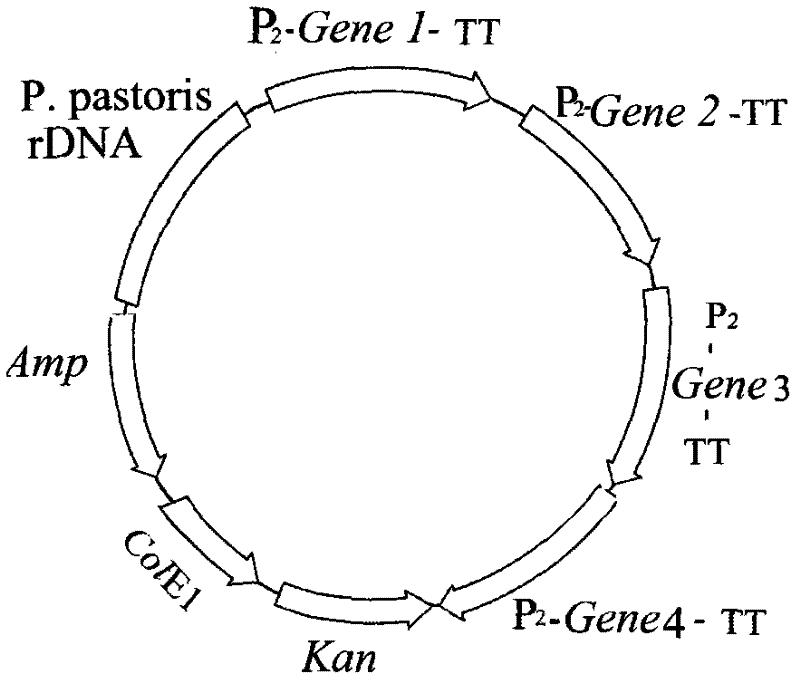
[0089] 2.1 Construction of Pichia pastoris engineering bacteria
[0090] 2.1.1 Construction of cloning vector
[0091] DNA synthesis contains AMP gene sequence, polyclonal linker and E. coli replication origin: base complementation is formed at both ends of the sequence. It is circularized by the action of DNA ligase to form a DNA cloning vector. The cloning vector was named pPCA.
[0092] 2.1.2 Acquiring genes
[0093] ①PCR amplification of isoamylase gene
[0094] Genomic DNA of Pseudomonas amyloliquefaciens was extracted, and primers (5'CG GATATC GCCATCAACAGCATGAGC3'; 5'CG GATATC GGCTACTTGGAGATCAACAG3') were used for PCR amplification. The PCR product was sequenced and analyzed with BLAST software provided by NCBI, which proved to be isoamylase gene sequence.
[0095] ②PCR amplification of α-amylase gene (including signal peptide) of Bacillus licheniformis
[0096] Genomic DNA of Bacillus licheniformis was extracted, and primers (5'Atgaaacaacaaaaacggct3'; 5'ctatctttga...
Embodiment 3
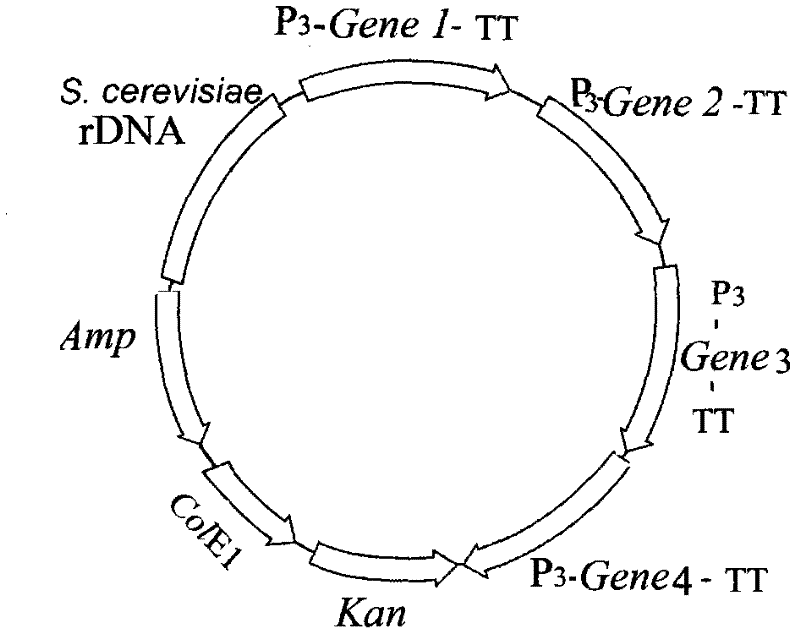
[0157] 3.1 Construction of Saccharomyces cerevisiae engineering bacteria
[0158] 3.1.1 Construction of cloning vector
[0159] DNA synthesis contains AMP gene sequence, polyclonal linker and E. coli replication origin: base complementation is formed at both ends of the sequence. It is circularized by the action of DNA ligase to form a DNA cloning vector. The cloning vector was named pSCA.
[0160] 3.1.2 Acquiring genes
[0161] ①PCR amplification of isoamylase gene
[0162] Genomic DNA of Pseudomonas amyloliquefaciens was extracted, and primers (5'CG GATATC GCCATCAACAGCATGAGC3'; 5'CG GATATC GGCTACTTGGAGATCAACAG3') were used for PCR amplification. The PCR product was sequenced and analyzed with BLAST software provided by NCBI, which proved to be isoamylase gene sequence.
[0163] ②PCR amplification of α-amylase gene (including signal peptide) of Bacillus licheniformis
[0164] Genomic DNA of Bacillus licheniformis was extracted, and primers (5'Atgaaacaacaaaaacggct3'; 5'c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com