Primer for quickly determining enterotoxigenic eschericha coli in feed sample and application for primer
A rapid detection technology for Escherichia coli, which is applied in the detection/testing of microorganisms, material stimulation analysis, biochemical equipment and methods, etc., to achieve the effects of high amplification efficiency, easy operation, and simplified detection procedures
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
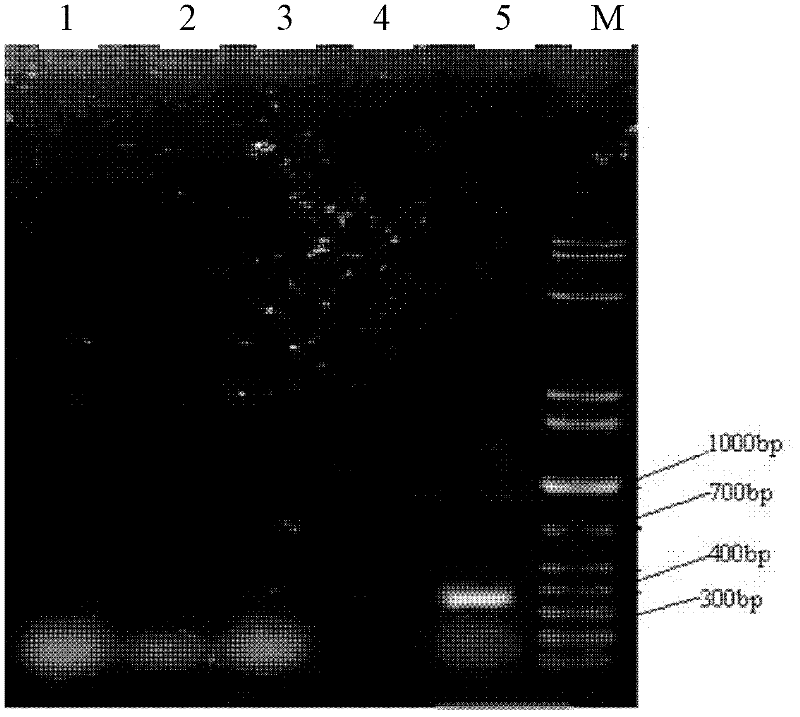
[0039] Example 1: Rapid qualitative detection of enterotoxic Escherichia coli K88 in predicted feed samples
[0040] This embodiment uses the gel recovery product of the target amplified fragment as a positive control; Sterile water is used as a negative control; In addition, Enterococcus faecalis, Staphylococcus aureus, and Salmonella are used as samples to carry out.
[0041] A. Preparation method of template DNA
[0042] (1) Take a certain amount of sample to be tested (bacteria liquid is generally 5ml, feed sample is 1000mg), and quickly grind it into powder under liquid nitrogen freezing; put the powder into a 2ml centrifuge tube, and then add 1MTris.Cl buffer 750ul (2) Place the centrifuge tube in a 65°C water bath for 1 hour, and mix gently during the water bath; take out the centrifuge tube, and add an equal volume of phenol-chloroform (V / V=1:1) solution to each tube 700ul, mix well and centrifuge at 10000rpm for 10 minutes; (3) transfer the supernatant to another cen...
Embodiment 2
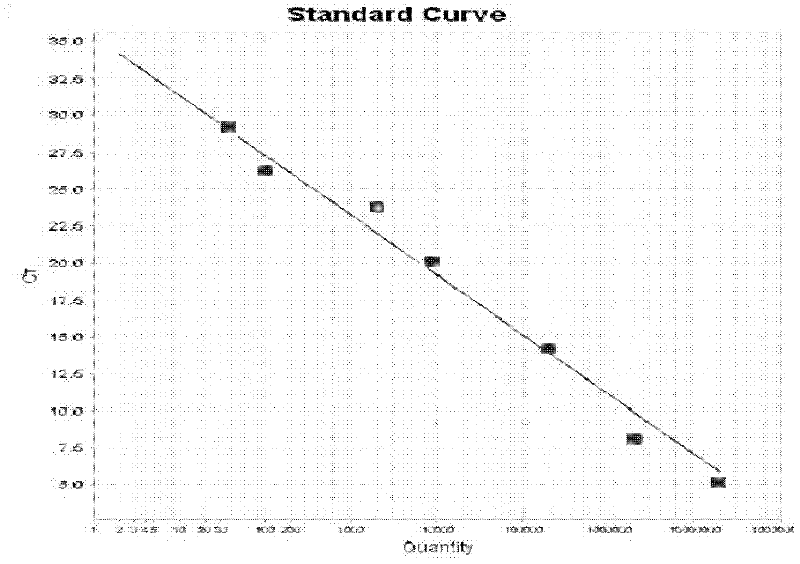
[0049] Example 2: Prediction of the preparation of the external standard of enterotoxic E. coli K88 in the feed sample and the drawing of the standard curve
[0050] After making a 10-fold serial dilution of enterotoxic Escherichia coli K88 with a known concentration, the concentration of the prepared bacteria was 10 7 -1CFU / ml according to
[0051] The method in embodiment 1 extracts bacterial DNA, and real-time fluorescent PCR detects, and the steps are as follows:
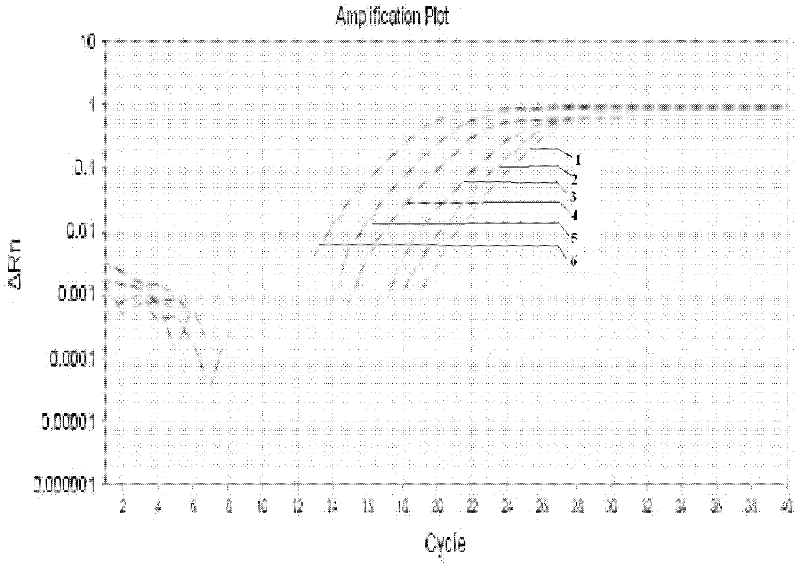
[0052] A. Fluorescent quantitative PCR
[0053] Reaction system 25.0 μL, including: 2×SuperReal PreMix mother solution 12.5 μL, upstream and downstream primers DCf and DCr 0.5 μL (20 μmol / L), DNA template 2.0 μL, 50×ROX Reference Dye 0.5 μL, RNase-free ddH 2 0 to 25 μL system;
[0054] Fluorescence quantitative PCR reaction parameters: pre-denaturation at 95°C for 10 minutes; denaturation at 94°C for 5s, annealing at 60°C for 15s, 40 cycles; storage at 4°C; repeat 3 times for each sample;
[0055] B. Prepara...
Embodiment 3
[0060] Embodiment 3: accuracy, stability and repeatability experiment
[0061] Using samples of the same concentration as templates, the methods in the national standard GB / T 4789.38-2008 and the method in Example 2 were used to detect the samples, and the two methods were compared. In order to evaluate the repeatability and stability of the test, the same batch was repeated twice for fluorescence quantitative reaction. For each group of test samples, 5 gradient reaction tubes were made, and the template concentration was divided into 2×10 7 , 2×10 6 , 2×10 5 , 2×10 4 , 2×10 3 CFU / μL. The results showed that the coefficients of variation of all dilutions were small (CV<5%), indicating that the inter-batch differences were small, and the fluorescent quantitative PCR reaction system established in this experiment had better intra-batch repeatability and stability (Table 1).
[0062] Substituting the Ct value shown in Table 1 into the standard curve in Example 2, the obtain...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com