Polymerase chain reaction (PCR) primer for cloning Actin gene of phalaris arundinacea linn and method
A technology of thorn grass and gene, which is applied in the field of molecular biology to achieve the effects of improving efficiency, shortening breeding years, and speeding up the breeding process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
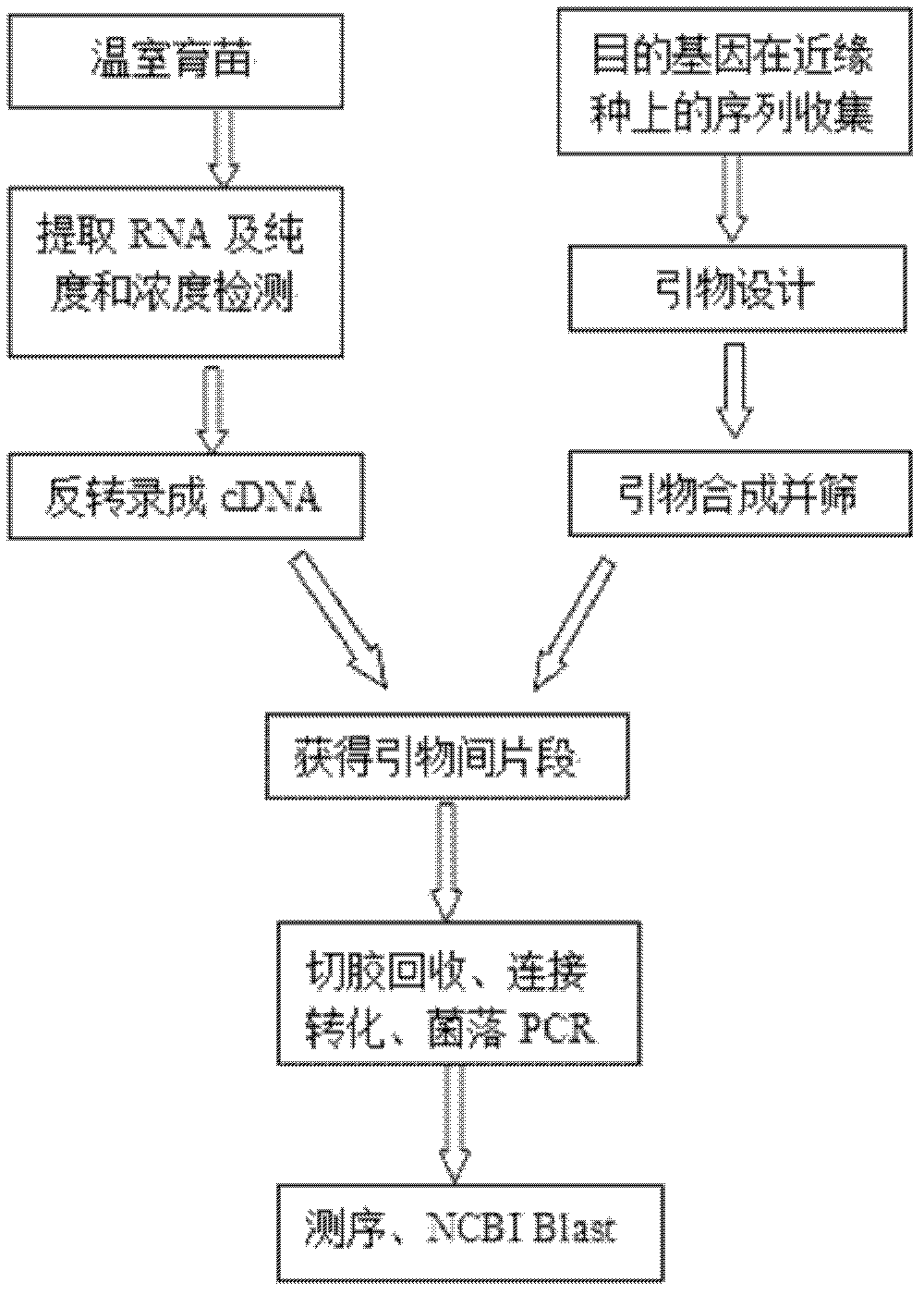
[0036] 1. Preparation of test materials
[0037] Pre-cooling and sterilizing the seeds of the weed grass, and planting them in plastic flowerpots after germination. The seedlings were grown for 30 days and used to extract RNA.
[0038] 2. Extraction of RNA
[0039] (1) Weigh about 0.2g of Lemongrass leaves, grind them into fine powder with liquid nitrogen, and divide them into two 1.5mL EP tubes (as fast as possible to avoid RNA degradation);
[0040] (2) Add 1mL TRIzol (Total RNA Extraction Reagent) to each tube, shake vigorously to make it evenly mixed, and leave it at room temperature for 5min (note that the amount of sample should not exceed 10% of TRIzol);
[0041] (3) When the content of protein, fat and polysaccharide in the sample is high, centrifuge at 12000rpm for 10min at 4°C, and suck the supernatant into a clean 1.5mL EP tube;
[0042] (4) Add 0.2mL chloroform to each tube, vibrate vigorously for 15 seconds, place at room temperature for 2-3min, and centrifuge at...
Embodiment 2
[0070] According to the cDNA sequences of the Actin genes of plants closely related to S. chinensis that have been registered on GenBank, they mainly include the sequences of rice, maize, stargrass, ryegrass and Leymus chinensis. The conserved regions of these cDNA sequences were analyzed with DNAMAN software, the specificity and other indicators of the primers were checked with Premier Primer 5.0, primers were designed, and primers were synthesized by Shanghai Sangong. Screen the primer pairs with good specificity among the designed primers and continue to do recovery conversion and linking experiments. The specific primers screened by the present invention are as follows:
[0071] Forward primer: 5′GACCGAGCGTGGTTACTC 3′
[0072] Reverse primer: 5′GCACCTCCAATCCAGACA 3′
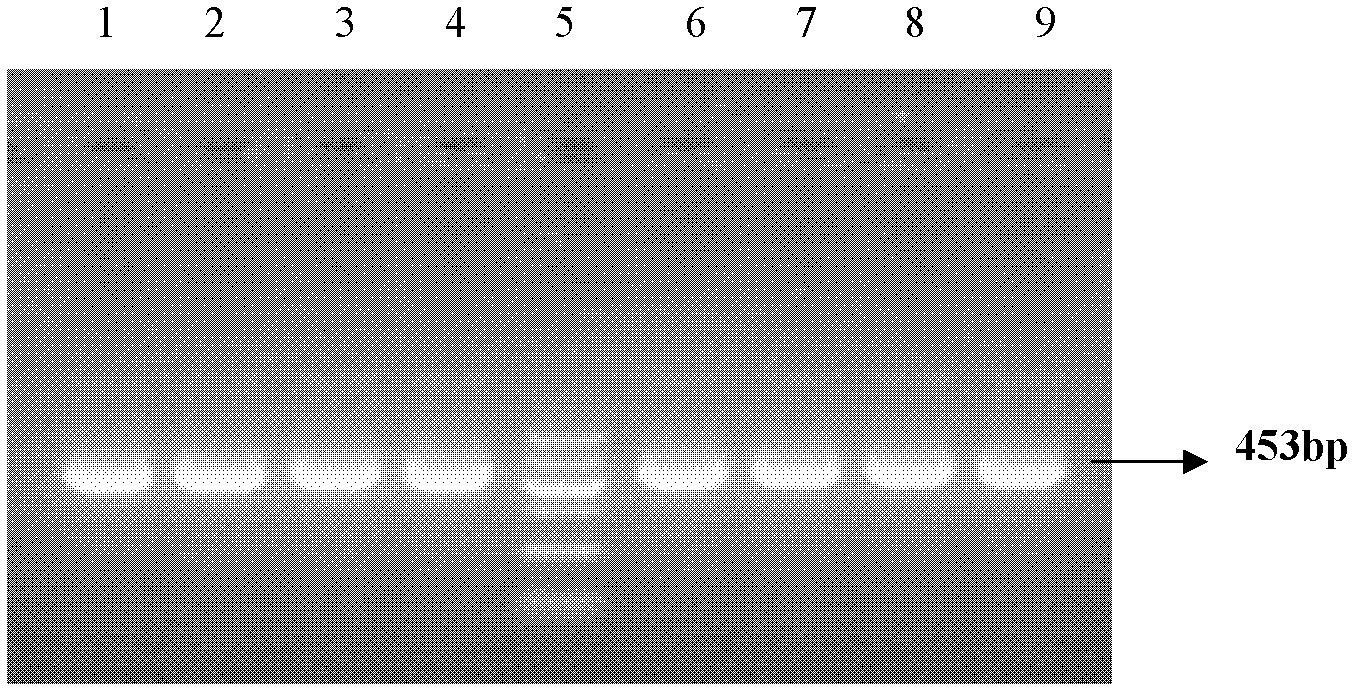
[0073] The cDNA obtained by reverse transcription in Example 1 was used as a template, and the above-mentioned primer sequence was used as a PCR reaction primer to carry out PCR amplification. The amplifica...
Embodiment 3
[0078] 1. The PCR gel electrophoresis band in Example 2 is subjected to gel cutting to recover the target fragment.
[0079] (1) After the DNA fragments to be recovered are separated by electrophoresis, cut out the required DNA fragments from the agarose gel, put them in a 1.5mL centrifuge tube, and centrifuge at a low speed.
[0080] (2) Add 3 times the volume of sol solution, 50°C metal bath (heating block) sol, and gently flip the centrifuge tube until the agarose gel is completely dissolved.
[0081] (3) After the gel solution is cooled to room temperature, add it to an adsorption column, centrifuge at 12,000 rpm for 30 seconds, discard the waste liquid in the collection tube, and put the adsorption column back into the collection tube.
[0082] (4) Add 700 μl of rinse solution to the adsorption column, centrifuge at 12000 rpm for 30 seconds, discard the waste liquid in the collection tube, and put the adsorption column back into the collection tube.
[0083] (5) Add 500 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com