Gene chip for detecting porcine respiratory disease complex virus and method for detecting porcine respiratory disease complex virus
A technology for porcine respiratory diseases and syndrome virus, which is applied in the field of gene chip and detection, can solve the problems of difficult and difficult diagnosis, etc., to improve the efficiency of diagnosis, improve the speed and accuracy of diagnosis, and have good repeatability and specificity. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1: Design and preparation of probes:
[0041] 1. Sequence acquisition of oligonucleotide probes:
[0042] Download the NPS2 gene of porcine reproductive and respiratory syndrome virus (PRRSV), the gene between 990 and 1500 bp of the genome sequence of circovirus type II (PCV-2), and the gE gene and pK gene of pseudorabies (PRV) from the Genbank public database , HA, NA and M genes of swine influenza virus H1N1, HA and NA gene sequences of swine influenza virus H3N2;
[0043] 2. Probe design for oligonucleotide probes:
[0044] Import the above sequence into Primer Premier 5.0 software, extract the ORF sequences of all viruses, and perform global alignment, obtain the characteristic segment of a specified virus according to the alignment map, design specific probes in the characteristic region, and the specific site should be as close as possible to the In the middle of the probe or slightly close to the 3' end of the probe, set the parameters, try to set the Tm ...
Embodiment 2
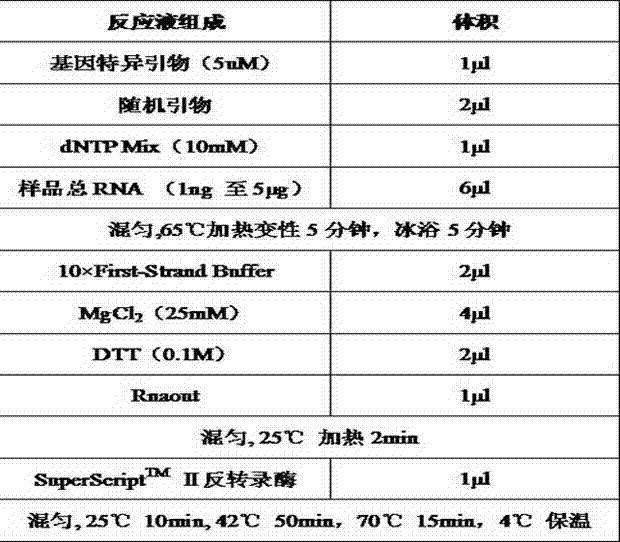
[0055] Example 2: Design and preparation of primers
[0056] 1. Sequence acquisition:
[0057] Download the NPS2 gene of porcine reproductive and respiratory syndrome virus (PRRSV), the gene between 990 and 1500 bp of the genome sequence of circovirus type II (PCV-2), and the gE gene and pK gene of pseudorabies (PRV) from the Genbank public database , HA, NA and M genes of swine influenza virus H1N1, HA and NA gene sequences of swine influenza virus H3N2;
[0058] 2. Design primers:
[0059] Import the above sequence into Primer Premier 5.0 software, set the parameter Tm value to 50°C-70°C, length 20bp±2bp, and then run the program.
[0060] 3. Primer selection:
[0061] From the output results, select primers with a Tm value of 50° C. to 70° C., a length of 20 bp±2 bp, and including the probe sequence used in the gene chip. Individual probes are manually adjusted, and the primers are appropriately increased or shortened by a few bases to make them contain probes and meet ...
Embodiment 3
[0066] Example 3: Gene Chip Preparation - Chip Spotting
[0067] 1. Dissolving the probe: Dissolve the probe synthesized in the above Example 1 in 50% DMSO, mix well with a shaker, and then centrifuge quickly to remove the liquid on the tube wall. After standing at room temperature for 1 hour, take 1 μL and dilute it with 50% DMSO to measure OD, measure the nucleic acid concentration of the probe, and then dilute the probe to a final concentration of 40 μM.
[0068] 2. Add plate: Add 5 μL 30-60 μM oligonucleotide aqueous solution to the 384-well plate, add 5 μL 2× gene chip sample solution, and mix thoroughly with a pipette.
[0069] 3. Spotting: use the automatic spotting instrument to spot the probes in the above-mentioned 384-well plate on the glass slide to form a designed microarray. In this embodiment, it is completed by using the crystal core microarray chip spotting system produced by Boao Biotechnology Co., Ltd. The specific operation steps are as follows: use the c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com