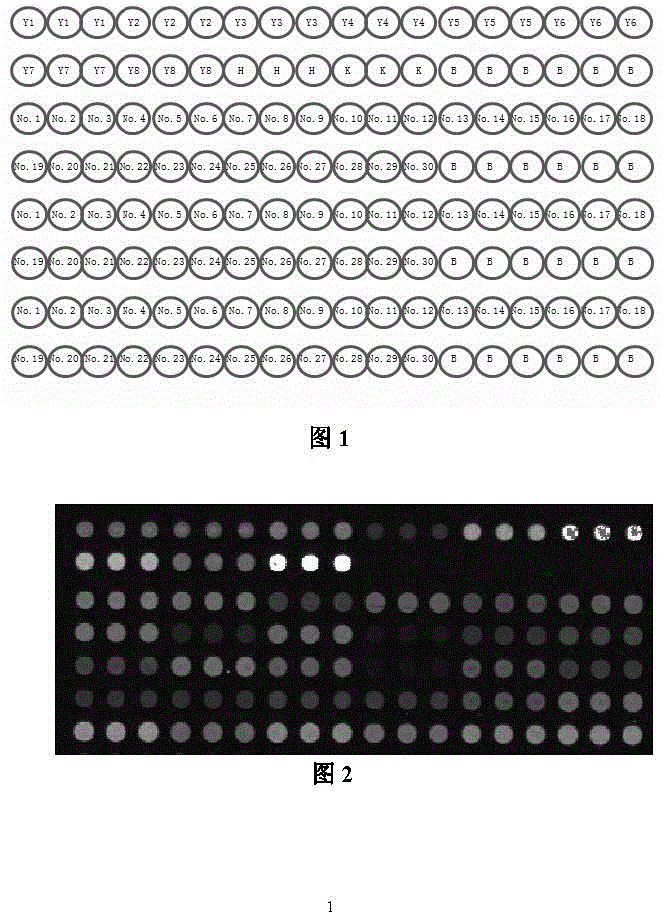
Echinococcus granulosus developmental-stage secretory protein expression gene chip
A technology of Echinococcus granulosus and gene chip, which is applied in protein nucleotide library, microbial determination/inspection, chemical library, etc. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Embodiment 1, the construction of cDNA library
[0028] The Creator SMART cDNA library construction kit provided by Clontech was used to construct the Eg cDNA library according to its instructions.
[0029] 1. Extraction of total RNA
[0030] The method of Chomezynski and Sacchi (1988) was used for preparation of total RNA and oligo-dT cellulose chromatography for poly(A)+ RNA (Sambrook et al., 1989).
[0031] Protoscoleum of Echinococcus granulosus was extracted from fresh goat liver tissue suffering from cystic echinococcosis, and frozen in liquid nitrogen. Take out 100 mg of tissue, grind it into powder in liquid nitrogen, add 1 ml Trizol solution for grinding, place the grinding solution at room temperature for 5 minutes, transfer it to a 2 mL glass tube (treated with DEPC), homogenize it on ice for 20 times, and transfer it to 1.5 mL EP tube, then add 0.2ml of chloroform per 1ml of Trizol solution, cap the centrifuge tube tightly, shake the centrifuge tube vigorous...
Embodiment 2
[0063] Example 2, cDNA library sequencing
[0064] 1. Colony PCR identification
[0065] (1) Randomly select 40,000 single colonies from the cDNA library and culture them overnight in 5mL LB medium. Take 1 μl of bacterial solution from each tube as a template, and perform PCR amplification with M13 universal primers (forward primer is 5'-CCCAGTCACGACGTTGTAAAACG-3', reverse primer is 5'-ACGGATAATTTCACACAGG-3') to confirm Whether the clone has the target fragment inserted and whether it is a single product.
[0066] (2) Establish the PCR reaction mixture (MasterMix) according to the conditions in Table 2 (that is, the PCR reaction formula table in Table 2)
[0067] (3) Mix well and centrifuge briefly.
[0068] (4) Carry out PCR reaction, the reaction parameters are as follows: 94°C for 30 seconds for 1 cycle, (94°C for 30 seconds, 68°C for 3 minutes) for 35 cycles, 68°C for 5 minutes for 1 cycle.
[0069] (5) At the end of the cycle, take 5ul products respectively, and obser...
Embodiment 3
[0074] Example 3 Preparation of gene chip
[0075] 1. Re-amplification of new expressed sequence tags
[0076] Select the PCR products of SEQ ID No.1 to SEQ ID No.30 as templates, dilute them 100 times, and carry out PCR amplification (DNAengine PCR instrument-Bio-Rad, USA) respectively. The primer is the universal primer M13 (the forward primer is 5'- CCCAGTCACGACGTTGTAAAACG-3', the reverse primer is 5'-AGCGGATAATTTCACACAGG-3'), each 100 μl reaction system includes: 10×PCR buffer 10 μl; 25 mmol / L MgCl 2 7 μl; 1 μl each of forward and reverse primers of 100 ng / μl, 1 μl of 20 mmol / L dNTP, 3 U of Tag DNA Polymerase (Promega, USA) and about 10 ng of plasmid template. PCR amplification process: pre-denaturation at 94°C for 4 min, denaturation at 94°C for 30 s in each cycle, annealing at 58°C for 60 s, extension at 72°C for 120 s; after 36-40 cycles, extension at 72°C for 10 min.
[0077] 2. PCR product processing
[0078] Add 200 μl (2 times the volume) of ethanol or 100 μl of...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com