N-acylhomoserine lactonase QsdA-RH5 with substrate specificity and coding gene and application thereof
A technology of acyl homoserine lactone and coding gene, which is applied in the field of substrate-specific N-acyl homoserine lactonase QsdA-RH5 and its coding gene and application, which can solve the problems of normal regulation and lack of substrate specificity. Oneness and other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
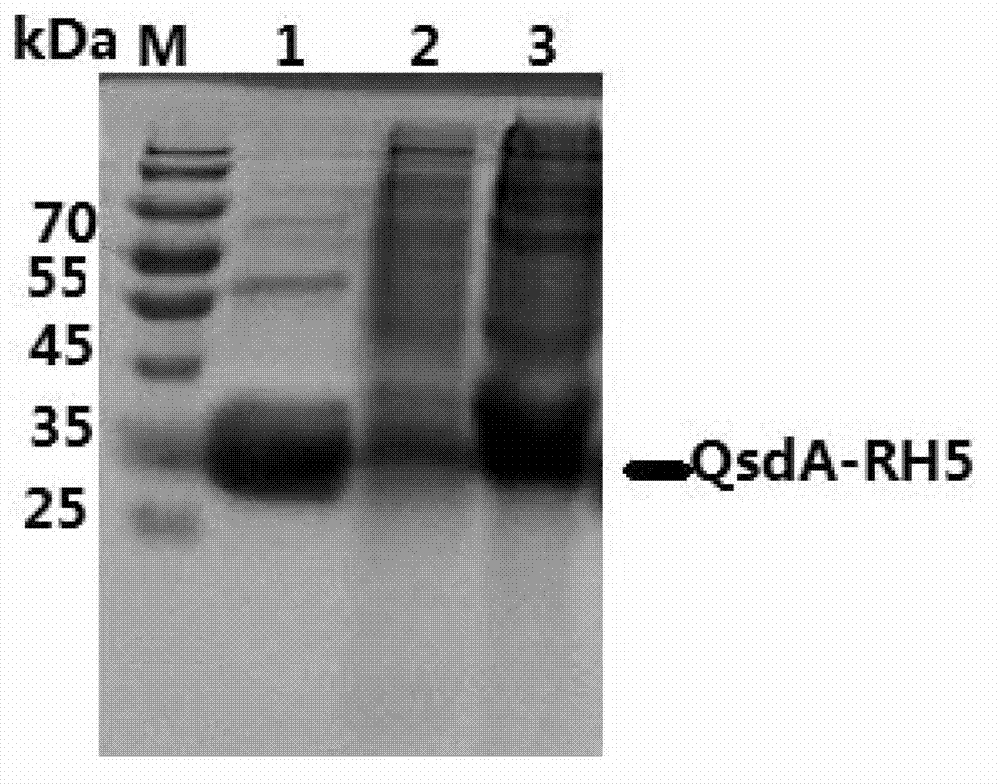
[0058] Embodiment 1, acquisition of Rhodococcus erythropolis QsdA-RH5 protein and its coding gene
[0059] Inoculate Rhodococcus erythropolis (Rhodococcus erythropolis) BFXJ-1 in LB liquid medium, incubate at 30°C for 24 hours, take 1ml of the bacterial liquid, and centrifuge at 10,000 rpm for 2 minutes to collect the bacterial cells, extract genomic DNA, and use primers Qs dA-F: 5′ -AGTTCAGTACAAACCGTTCGTG-3′ and Qs dA-R: 5′-TCAGCTCTCGAAGTACCG-3′ were amplified by PCR; PCR reaction conditions were 95°C for 5min, 94°C for 30sec, 55°C for 30sec, 72°C for 1min, 30 cycles, 72°C for 10min Perform agarose gel electrophoresis on the obtained PCR amplification product, recover and purify a band of about 1000 bp, connect it to the vector pEASY-T3, and sequence the obtained recombinant plasmid. The results show that the recombinant plasmid is contained in the vector pEASY-T3 The DNA shown in the sequence 2 of the sequence listing from the 4th to the 969th nucleotide at the 5' end was ...
Embodiment 2
[0061] Embodiment 2, the preparation of QsdA-RH5 protein
[0062] 1. Construction of recombinant expression vector
[0063] 1. Synthesize the DNA shown in Sequence 3 of the Sequence Listing, which contains protective bases and enzyme recognition sequences at both ends.
[0064] 2. Using the DNA synthesized in step 1 as a template, perform PCR amplification with a primer pair composed of QsdA 2-F and QsdA 2-R to obtain a PCR amplification product.
[0065] QsdA 2-F: 5′-CG GAATTC ATGAGTTCAGTACAAACCGTTCGTG-3' (the underlined base is the recognition sequence of EcoR I digestion);
[0066] QsdA 2-R: 5′-AA GCGGCCGC TCAGCTCTCGAAGTACCG-3' (the underlined base is the recognition sequence of Not I restriction enzyme).
[0067] 2. Digest the PCR amplified product of step 1 with restriction endonucleases EcoR I and Not I, and recover the digested product.
[0068] 3. Digest the vector pET-28a(+) with restriction endonucleases EcoR I and Not I to recover the vector backbone (about 5...
Embodiment 3
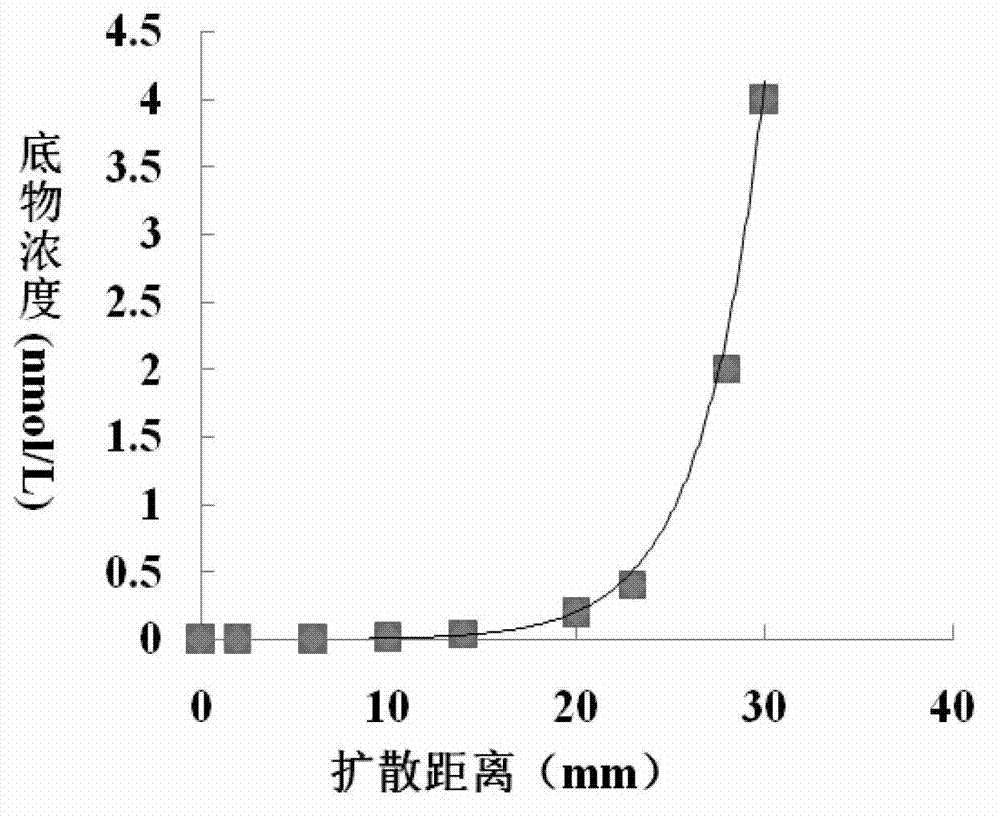
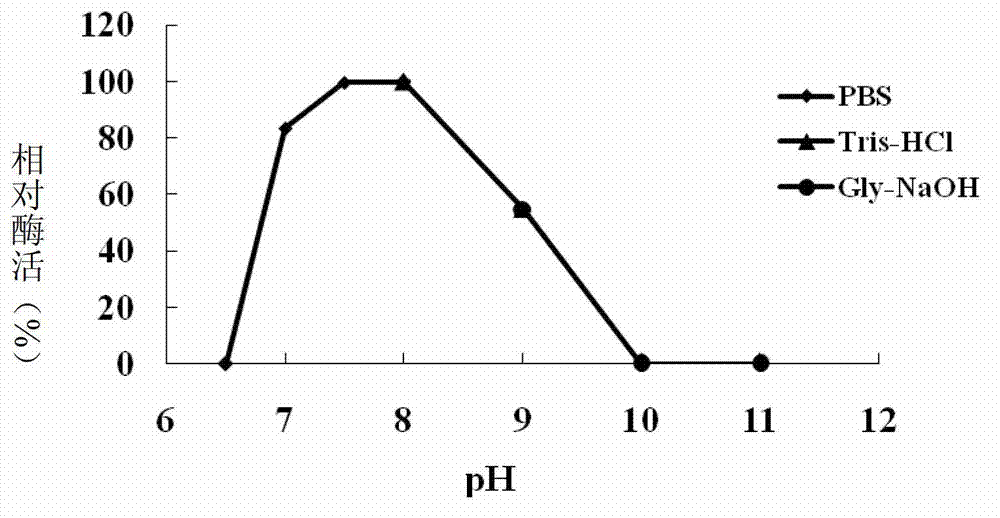
[0105] Example 3, Characterization of QsdA-RH5 protein as N-acyl homoserine lactonase
[0106] 1. Optimal pH
[0107] Method: Prepare the QsdA-RH5 protein solution obtained in step 3 of Example 2 with 0.1mol / L PBS buffer (pH8.0) to prepare a solution with a total protein concentration of 1.0mg / mL (as the solution to be tested), according to The method in step 4 of Example 2 was used to detect N-acyl homoserine lactonase activity. The difference was that the 0.1mol / L PBS buffer solution (pH8.0) in the reaction system used the following buffer solutions with different pH values:
[0108] 0.1mol / L McIlvaine buffer (citric acid) at pH 4.0, 5.0, 6.0;
[0109] 0.1mol / L PBS buffer (PBS) with pH 5.0, 6.0, 7.0, 7.5, 8.0;
[0110] Tris-HCl buffer (Tris-HCl) at pH 8.0, 8.5, 9.0;
[0111] 0.1mol / L glycine-NaOH buffer (Gly-NaOH) with pH 9.0, 10.0, 11.0.
[0112] When 0.1mol / L PBS buffer solution (pH8.0) is used in the reaction system, the enzyme activity of the solution to be tested is...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com