Primer and probe for detecting FMO3 gene A1034T mutation of chicken
A technology of A1034T and FMO3-, which is applied in the field of TaqMan MGB probes, can solve the problems that the detection results are easily affected by experimental conditions, cumbersome steps, and low sensitivity, and achieve reliable detection results, simple workflow, and high sensitivity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Embodiment 1 detects the design of specific primers and probes of chicken FMO3 gene A1034T mutation site
[0047] The invention obtains a pair of fluorescent quantitative PCR primers for detecting the A1034T mutation site of the chicken FMO3 gene and a TaqMan probe used in conjunction with the primers through repeated screening and verification.
[0048] Upstream primer FMO3 F: 5'-GTGCAGGACGACCTCGAT-3' (SEQ ID NO.1)
[0049] Downstream primer FMO3 R: 5'-CTCCATGAAAGGAAAGGAGTGAGA-3' (SEQ ID NO.2).
[0050] The upstream primer consists of 18 bases, corresponding to bases 1001-1018 of the chicken FMO3 gene cDNA sequence in GenBank (accession number: AJ431390); the downstream primer consists of 24 bases, corresponding to the 1043rd base of the chicken FMO3 gene cDNA sequence ~1066 base sequences. The length of the amplified fragment of the primer pair is 66bp.
[0051] The nucleotide sequence of the TaqMan fluorescent probe used in conjunction with the above specific prim...
Embodiment 2
[0055] Example 2 Chicken Anticoagulant Genomic DNA Extraction
[0056] This example contains a total of 50 Beijing oil chickens, of which 6 young chickens (8 weeks old) are used for the establishment of the TaqMan probe real-time fluorescent quantitative PCR detection method, and the other 44 hens are 40-week-old hens waiting for pure breeding. Choose to keep the hen. They were raised in the Yufa Breeding Chicken Farm of Beijing Academy of Agriculture and Forestry Sciences, blood was collected from the subwing vein, and anticoagulated with ACD (citric acid 0.48g, sodium citrate 1.32g, glucose 1.47g, added distilled water to 100ml).
[0057] Chicken Anticoagulant Genomic DNA Extraction Kit DP318 (Tiangen Biochemical Technology Co., Ltd., Beijing) was used to extract genomic DNA. The specific steps are as follows:
[0058] 1) Take out the sample, after melting, pipette 20μL into a 2mL centrifuge tube;
[0059] 2) Add 180 μL buffer GS, 20 μL proteinase K, and mix well;
[0060...
Embodiment 3
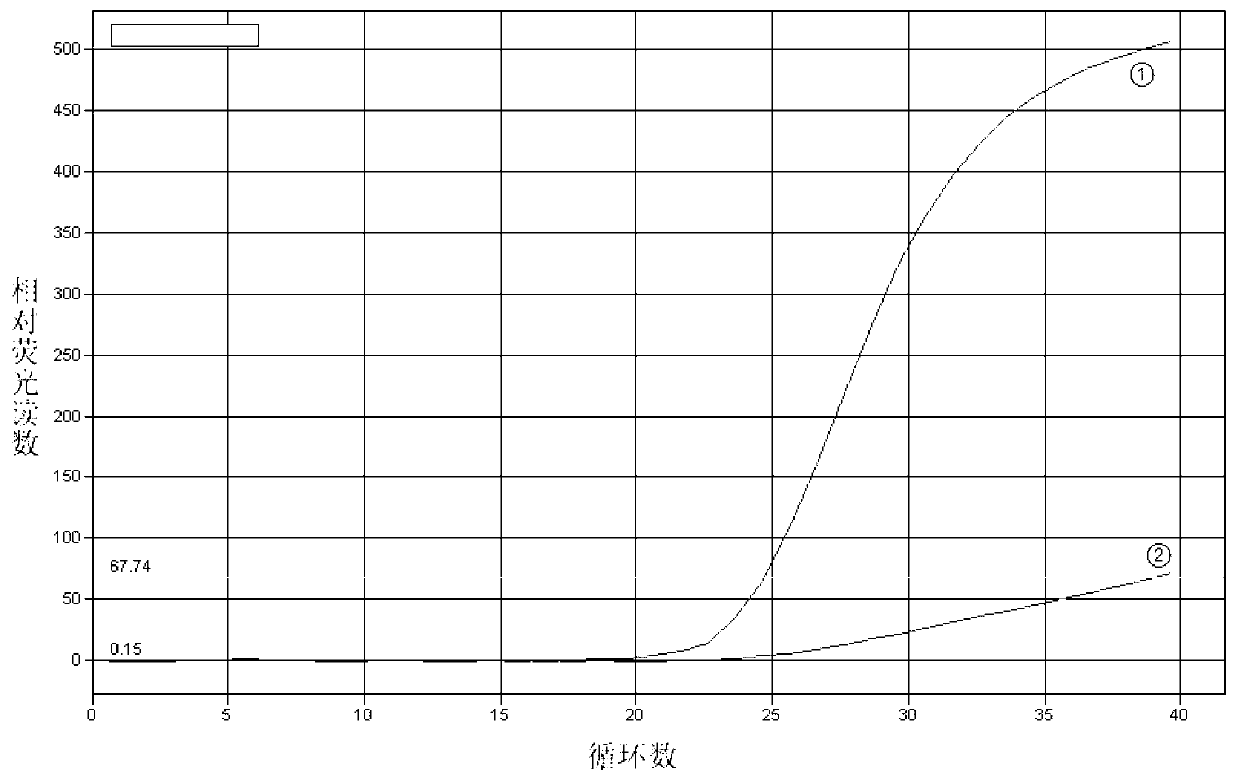
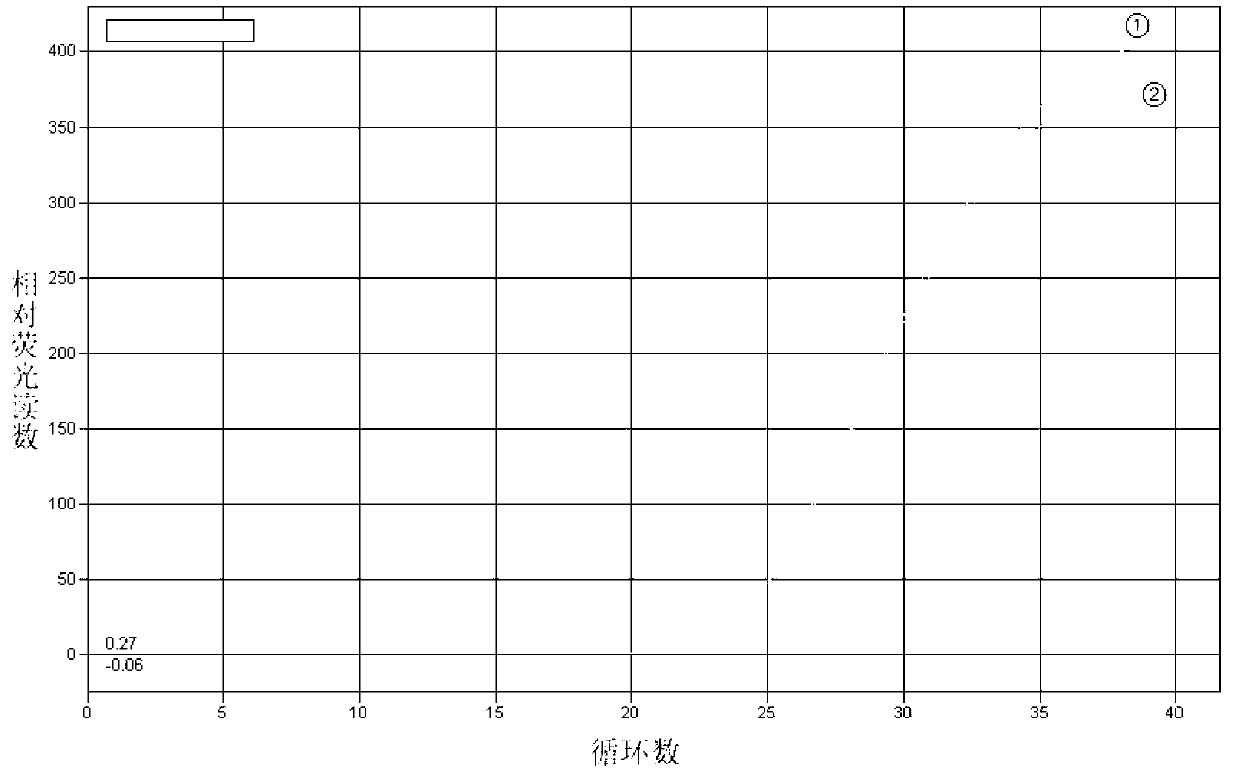
[0071] Example 3 Establishment of TaqMan probe real-time fluorescent quantitative PCR detection method for chicken FMO3 gene A1034T mutation
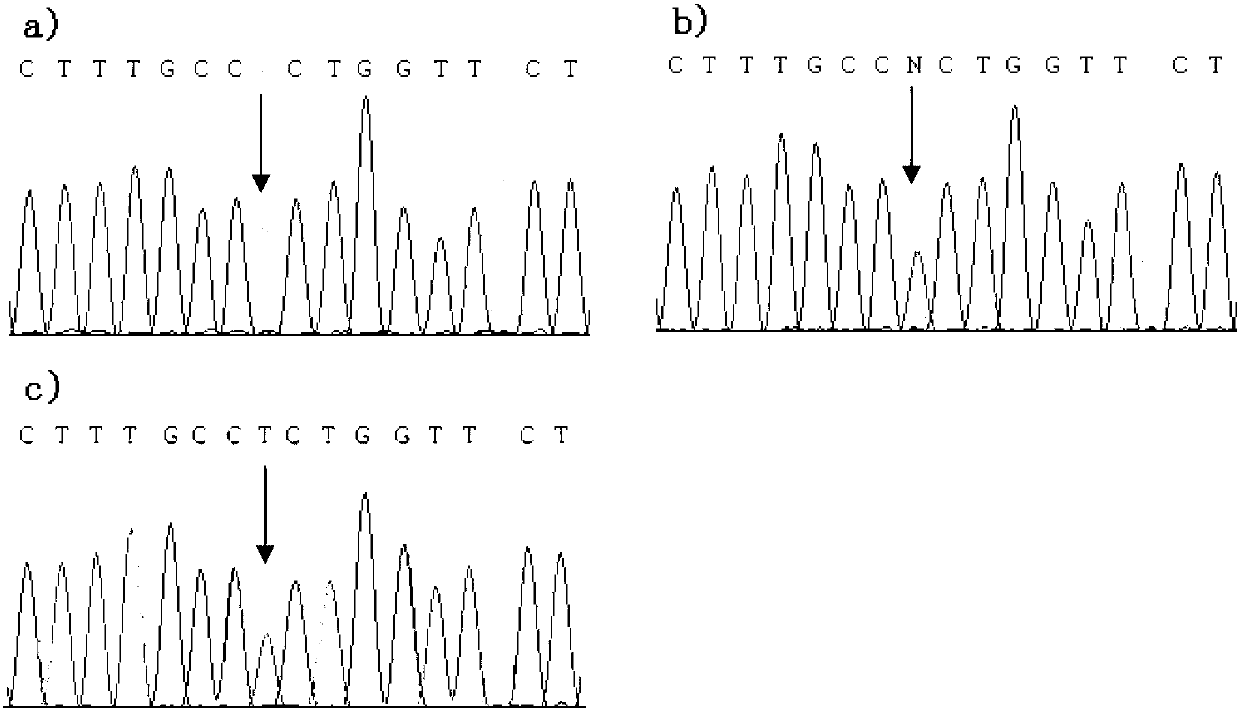
[0072] 1. Determination of sample DNA genotype
[0073] The DNA concentration of the 6 chickens obtained in Example 2 was detected again using NanoDrop2000 chromatography, and the DNA concentrations of chickens A, B, C, D, E, and F were respectively: 14.5ng / μL and 14.5ng / μL , 13.3ng / μL, 32.2ng / μL, 23.2ng / μL, 29.9ng / μL.
[0074]Using the genomic DNA of the above 6 chickens as a template, PCR amplification was performed using the following primer pair (Zhang Longchao, 2006). The upstream primer FMO_F1: 5'-CAGGGTGGTATCAAGCCTGT-3', the downstream primer FMO_R1: 5'-GATCCAAAGGACTGGACCAA-3' . The amplification reaction system is a 25 μL system, including: 2 μL (50 ng) of the genomic DNA template of 6 chickens obtained in Example 2, 0.5 μL of Taq enzyme (Takara), 2.5 μL of 10×buffer, 2.0 μL of dNTP, upstream and downstream primers (10 pmol / ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com