Primer set for real-time fluorescent quantitative PCR detection of swine influenza virus
A swine influenza virus and primer set technology, applied in the biological field, can solve the problems of high price, cumbersome probe design and synthesis, and achieve good specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0020] Experimental operation steps of the present invention are as follows:
[0021] 1. Extraction of viral RNA: Trizol reagent from Invitrogen Company was selected, and viral RNA was extracted according to the instructions of the reagent.
[0022] 2. Preparation of cDNA: Use the RNA extracted in step 1 to prepare cDNA using the TIANScript cDNA First Strand Synthesis Kit from Tiangen Company.
[0023] 3. Real-time PCR: The selected Real-time PCR kit is from TAKARA company Premix Ex Taq TM II (Perfect Real Time), each sample set 3 replicates. The reaction system and its components are shown in Table 2.
[0024] Table 2 Real-time PCR reaction system and its components
[0025]
[0026] Amplification conditions used Premix Ex Taq TM Standard amplification conditions provided by the II (Perfect Real Time) kit, such as 95°C for 30 seconds; 95°C for 15 seconds; 60°C for 30 seconds for 40 cycles; melting.
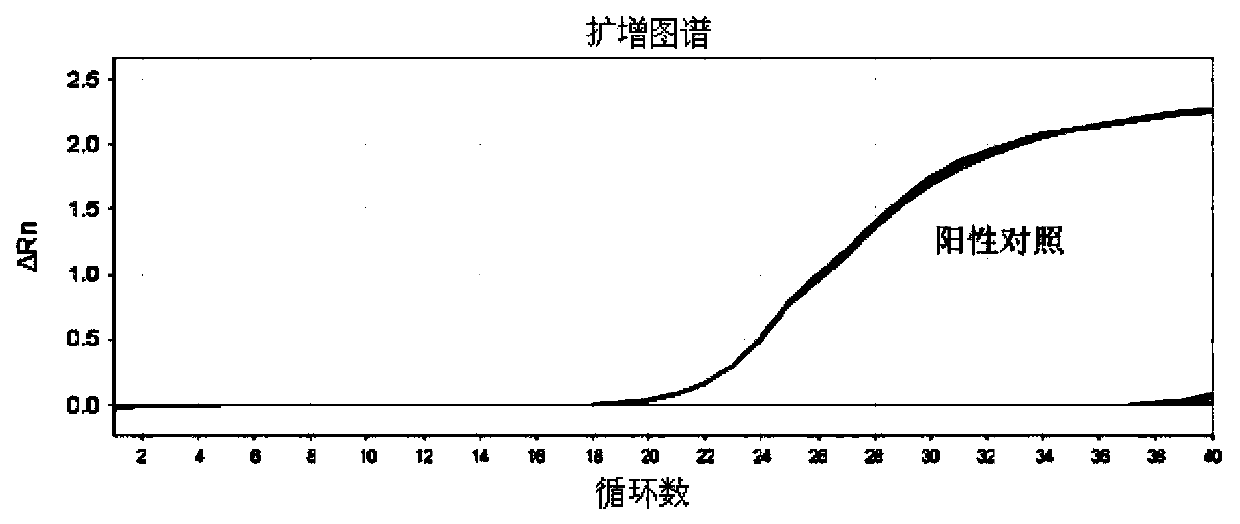
[0027] When performing Real-time PCR detection, when the CT va...
Embodiment 2
[0029] Example 2 Primer Sensitivity Detection
[0030] 1. Sample: H1N1SIV (10 8 EID 50 / ml) or H3N2SIV (10 8 EID 50 / ml) as a reference, and nuclease-free water as a negative control. H1N1 SIV is used to detect the sensitivity of SIV-M, SIV-H1 and SIV-N1 primers. H3N2 SIV is used to detect the sensitivity of SIV-M, SIV-H3 and SIV-N2 primers. Both H1N1 SIV and H3N2 SIV were isolated from pigs that clinically showed the main symptoms of SIV infection, and their subtypes were determined by the current general RT-PCR typing detection method.
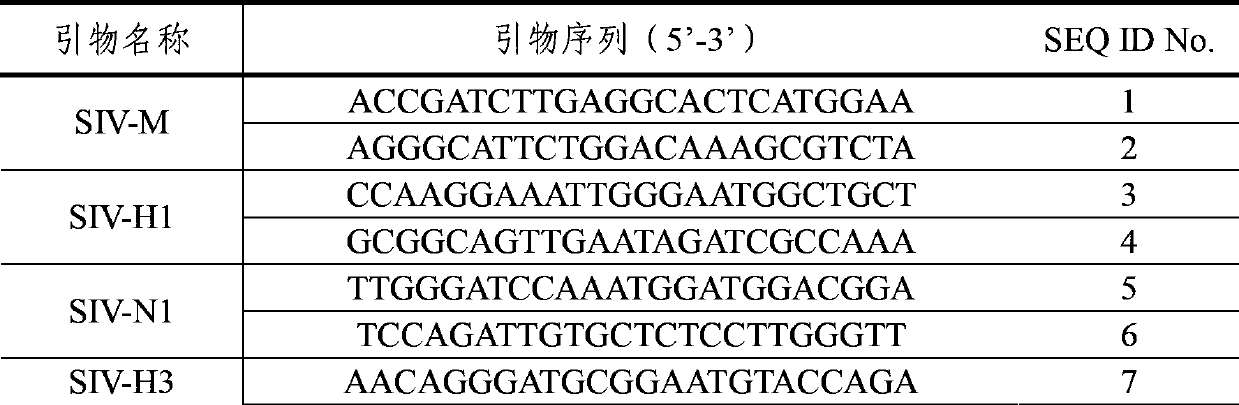
[0031] For SIV-Taq primers, see "A Real-time Fluorescent RT-PCR Detection Kit for Swine Influenza Virus" (Patent No.: 200910039407), which is a general primer for detecting swine influenza virus and a reference primer for the general primer SIV-M in this example , which is used to compare the sensitivity difference between the universal primer provided in the present invention and the universal primer in the existing invention.
[003...
Embodiment 3
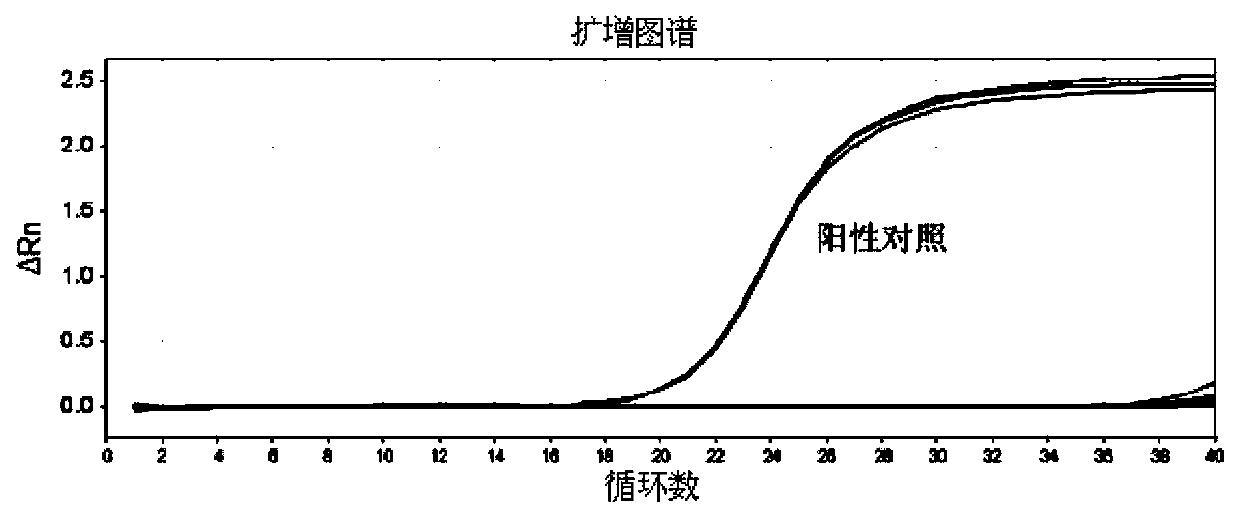
[0044] Example 3 Primer Specific Detection
[0045] 1. Samples: H9N2 avian influenza virus; common seasonal H1N1 influenza virus cDNA; 2009 pandemic H1N1 influenza virus cDNA. The subtypes of the above influenza virus samples were all determined by the current general RT-PCR typing detection method. H1N1 SIV and H3N2 SIV are the same as in Example 2.
[0046] H1N1 SIV is the positive control for the specific detection of SIV-H1 and SIV-N1 primers; H3N2 SIV is the positive control for the specific detection of SIV-H3 and SIV-N2 primers. The negative control was nuclease-free water.
[0047] 2. Operation steps:
[0048] 1) Extraction of viral RNA.
[0049] 2) Preparation of cDNA.
[0050] 3) Real-time PCR.
[0051] 3. Test results
[0052] Utilize the SIV-H1 and SIV-N1 primers provided by the present invention, and use the cDNA of common seasonal H1N1 influenza virus, 2009 pandemic H1N1 influenza virus, H9N2 avian influenza virus and H3N2 SIV as templates to detect the sp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com