Enhancing method for polymerase chain reaction
A technology of chain reaction and polymerase, applied in the biological field, can solve the problems of non-specific amplification, poor effect of template enhancement with high GC content, increased product production, etc., and achieve the effect of eliminating inhibition
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Example 1: Polymerase Chain Reaction (PCR) for simple linear templates L and G.
[0028] The template DNA sequence G is a template with high GC content and a thrombin protein recognition interval, and L is a random single-stranded DNA sequence. 100 μL reaction system contains 50 μL Taq enzyme PCR premix buffer, template L or G (600 or 6×10 7molecule), bovine thrombin protein (TB) 2.5μg / mL, forward primer (FP) and reverse primer (RP) 50pmol (Table 1). The PCR reaction conditions were, after pre-melting at 95°C for 30s, 33 rounds of cyclic amplification (denaturation at 94°C for 30s, annealing at 56°C for 30s, extension at 72°C for 30s). The products of the PCR reactions were kept at 4°C until analyzed by gel electrophoresis.
[0029] PCR products were separated by 10% non-denaturing polyacrylamide vertical gel electrophoresis (voltage 150V, time 50min) ( figure 1 ), After staining with gold nucleic acid dye for 10 minutes, the bands were analyzed by taking pictures ...
Embodiment 2
[0032] Example 2: Polymerase Chain Reaction of Random Single-Stranded DNA Libraries.
[0033] In a 50 μL reaction system, 25 μL PCR premix buffer containing Taq enzyme, 30 to 3×10 5 A single-stranded DNA library molecule, bovine thrombin protein 25μg / mL, forward primer (Pool-FP) and reverse primer (Pool-RP) 25pmol (Table 1). The polymerase chain reaction conditions were, after pre-melting at 95°C for 30s, 33 rounds of cyclic amplification (denaturation at 94°C for 30s, annealing at 56°C for 30s, extension at 72°C for 30s). The products of the PCR reactions were kept at 4°C until analyzed by gel electrophoresis.
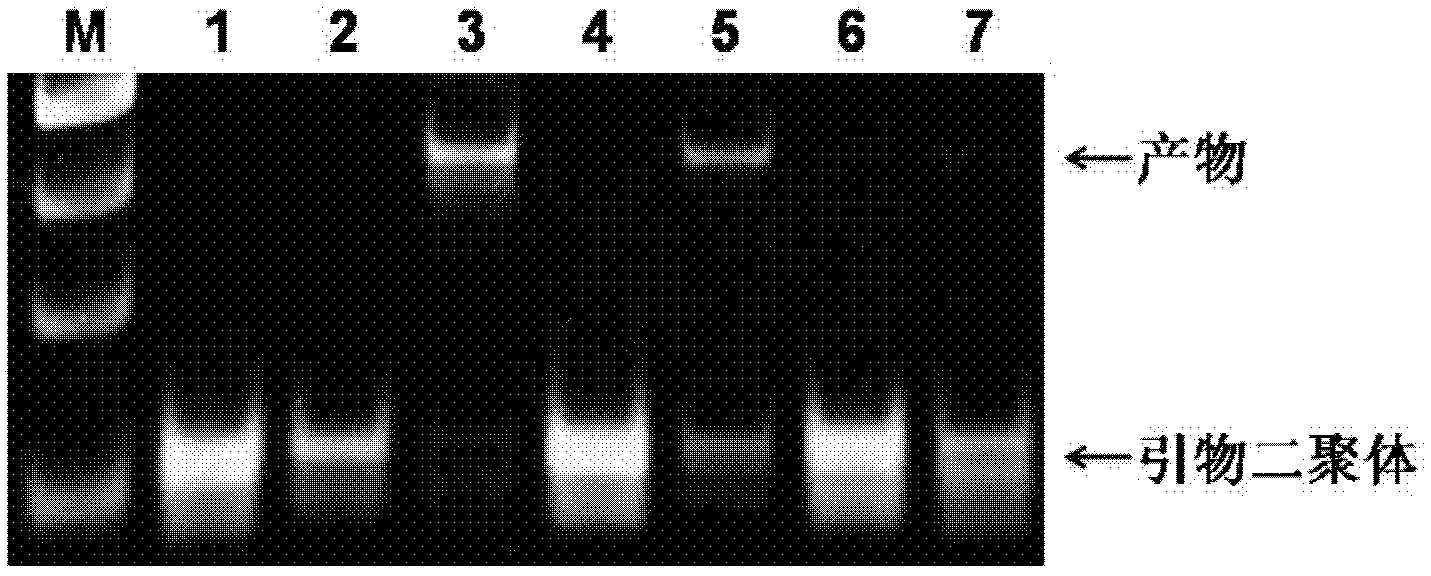
[0034] figure 2 It is a graph showing that bovine thrombin protein (TB) improves the amplification efficiency of a random single-stranded DNA library under low-concentration conditions. The initial template concentration was 3000 DNA molecules (lanes 2, 3), 300 DNA molecules (lanes 4, 5), and 30 DNA molecules (lanes 6, 7). The sample in lane 1 is a negative contr...
Embodiment 3
[0036] Example 3: Real-time quantitative PCR of a random single-stranded DNA library.
[0037] 20 μL reaction system contains 10 μL PCR premix buffer containing SYBR Green dye, 1.2×10 4 Single-stranded DNA library molecules, bovine thrombin protein (64 μg / mL), forward primer (Pool-FP) and reverse primer (Pool-RP) each 10 pmol (Table 1). The PCR reaction conditions were, after pre-melting at 95°C for 30s, 40 rounds of cyclic amplification (denaturation at 95°C for 20s, annealing at 56°C for 30s, extension at 72°C for 30s). The annealing temperature range is 55°C-95°C.
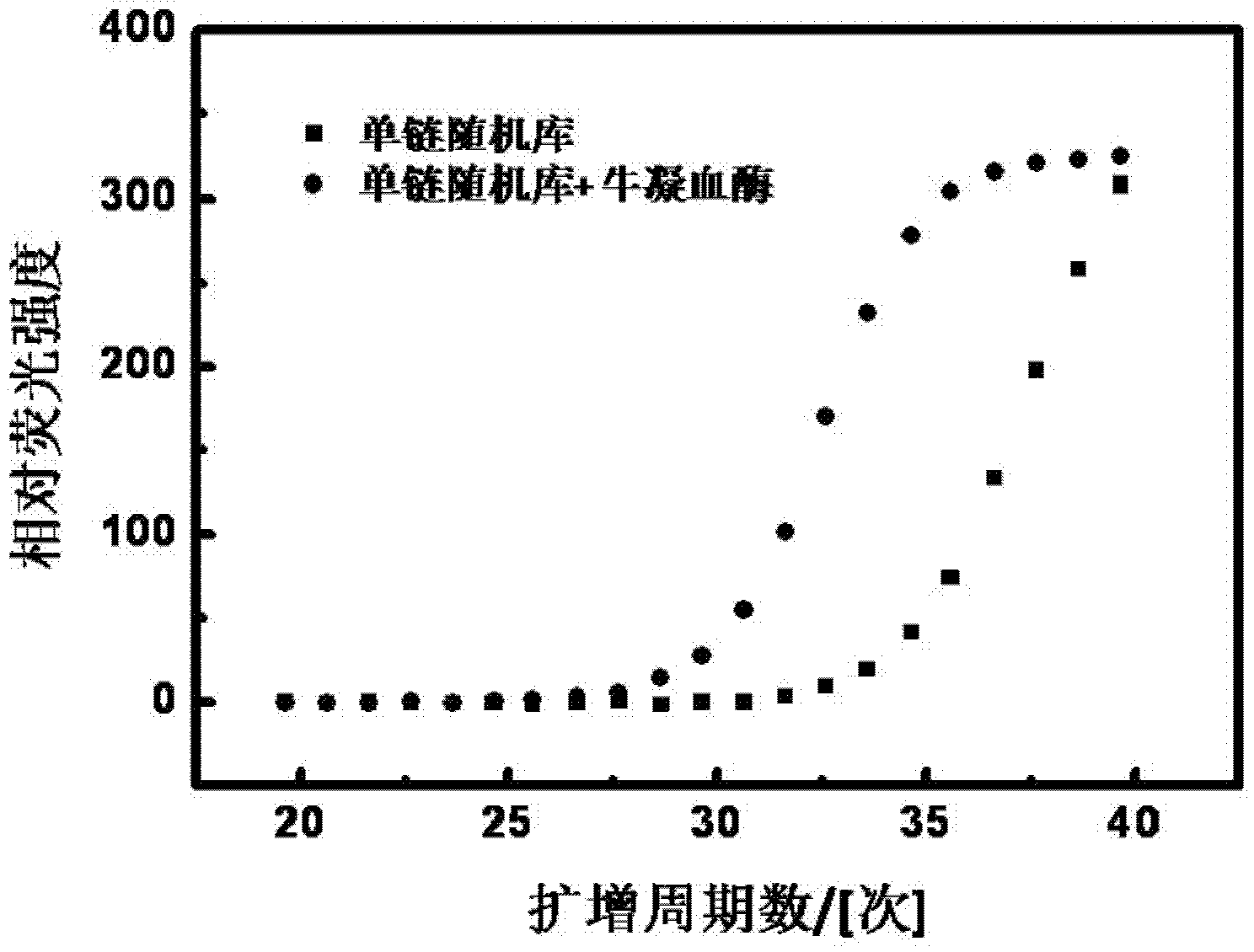
[0038] image 3 It is a graph showing that the addition of bovine thrombin protein (TB) to real-time quantitative PCR improves the amplification efficiency of a single-stranded random DNA library. 20 μL reaction system contains 1.2×10 4 A single-stranded DNA sequence was added at a concentration of 64 μg / mL of TB.
[0039] Figure 4 is a graph showing the real-time quantitative PCR annealing temperature pr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com