Method for constructing industrial saccharomyces cerevisiae engineering bacteria by integrating metabolism and rearrangement of gene
A technology of Saccharomyces cerevisiae and metabolic engineering, applied in the direction of microorganism-based methods, biochemical equipment and methods, fungi, etc., can solve the problems of difficult to obtain excellent strains with comprehensive performance, degradation and decay of key properties of production strains, and achieve reduction Energy consumption, high alcohol resistance, and low by-product effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1: Acquisition of Industrial Saccharomyces cerevisiae Glycerol Metabolism Engineering Strains
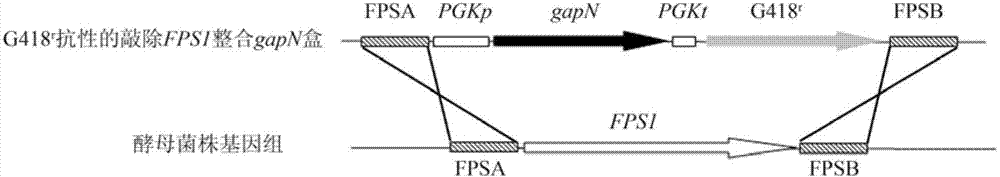
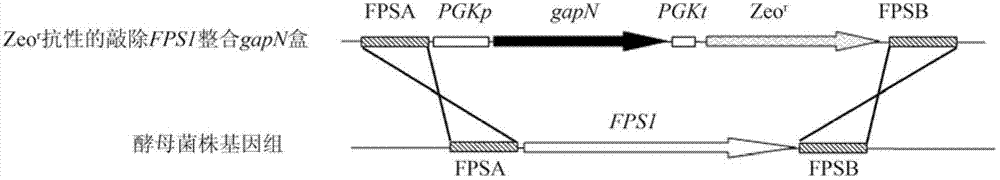
[0027] 1. Obtaining the haploid starting strain
[0028] After the industrial Saccharomyces cerevisiae Z87 was activated by YPD at 30°C, it was transferred into the sporulation medium and cultured at 26°C for 3-7 days. When the ascospores were observed under a microscope, the bacterial cells were collected, washed twice with normal saline, and then added with 700 μL Tris-HCl (pH8.0, 0.01 mol / L), 200 μL 100 mg / mL helicase solution and 100 μL 0.1 mol / L mercaptoethanol, Incubate at 30°C for 16 hours at 120r / min to rupture the ascus wall and release spores. Treat the vegetative cells at 58°C for 15 minutes to kill the vegetative cells, collect the spores by centrifugation, spread them on a YPD plate, incubate at 30°C for 2-3 days, pick a single colony, and verify the sporulation after activation on the YPD slant. Somatic strains.
[0029]The strain to be tested and the...
Embodiment 2
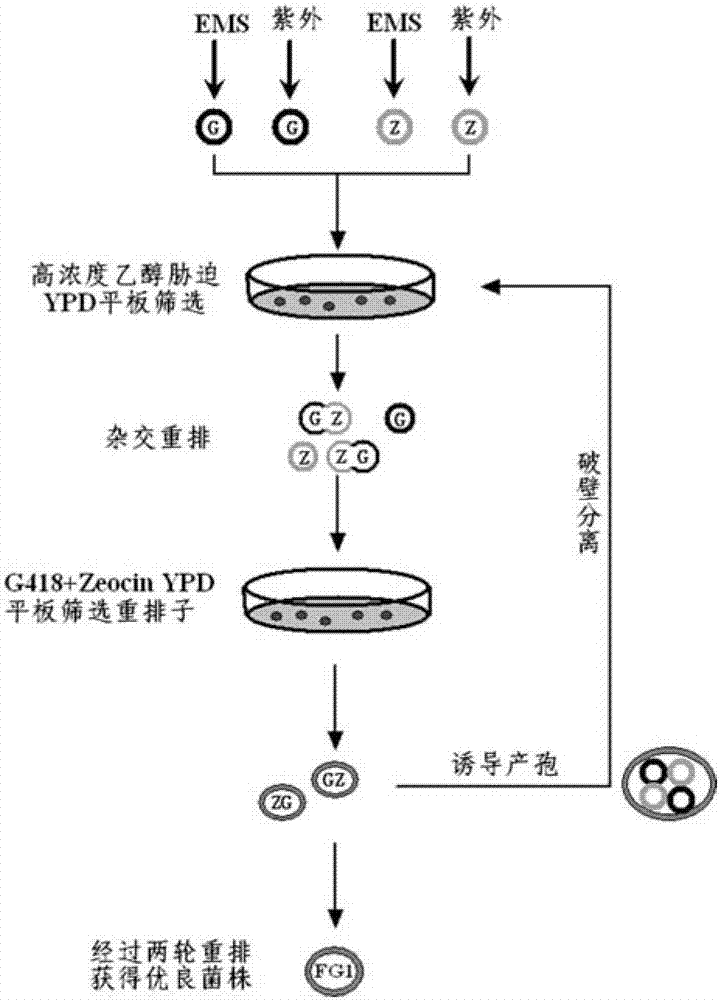
[0037] Embodiment 2: Implement whole genome rearrangement on genetically engineered strains
[0038] Such as image 3 The shown process implements whole genome rearrangement, and the specific steps are as follows:
[0039] 1. 1% (v / v) EMS mutagen was used to treat genetically engineered haploid strains YFG1 (MAT a, fps1Δ::PGKp-gapN) and YFG2 (MATα, fps1Δ::PGKp-gapN) for 30-120 minutes, Add 5% (w / v) sodium thiosulfate to remove EMS contamination every 30 minutes, collect bacteria by centrifugation at 4000rpm for 5 minutes, wash twice with normal saline, and apply gradient dilution to a YPD plate containing 8% (v / v) ethanol, 30 Cultivate for 3 days at ℃, and pick a vigorously growing single colony;
[0040] 2. Under the ultraviolet lamp with a distance of 40cm and a power of 15w, the genetically engineered haploid strains YFG1 (MAT a, fps1Δ::PGKp-gapN) and YFG2 (MATα, fps1Δ::PGKp-gapN) were subjected to ultraviolet mutagenesis , Sampling every 1min, diluted and coated with 8%...
Embodiment 3
[0044] Embodiment 3: Determination of production performance of engineering strains
[0045] 1. Growth determination under high-concentration ethanol stress conditions
[0046] The starting strain Z87, the control strain Y12, the genetically engineered strain YFG12, and the genetically engineered rearrangement FG1 were cultured in YPD liquid medium containing 0% and 10% (v / v) ethanol at 30°C, and samples were taken at different times to determine the dry weight of the strains. The maximum specific growth rate of the strain. Such as Figure 4 As shown, under the YPD culture condition containing 0% ethanol, there was no significant difference in the growth of the four strains, and the lag period was about 0.4h; under the YPD culture condition containing 10% (v / v) ethanol, the four strains lagged The average growth period was extended to about 10 hours, and the genetic engineering rearrangement FG1 grew the fastest, and the maximum specific growth rate was 11.4% higher than tha...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com