Highly specific gene segment of Cronobacter spp. and application thereof
A Cronobacter, high specificity technology, applied in the field of molecular biology, can solve the problems of poor specificity, low specificity and large sequence difference of 16S rDNA gene, and achieves low false positive rate, high specificity and high specificity. sexual effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Example 1: Acquisition of Specific Gene Fragments
[0042] 1. Using 16S rDNA universal primers:
[0043] F341: 5'-CCTACGGGAGGCAGCAG-3' (SEQ ID NO. 6),
[0044] R518: 5'-ATTACCGCGGCTGCTGG-3' (SEQ ID NO. 7);
[0045] Using Cronobacter genomic DNA as a template, the target sequence was amplified. PCR reaction system (50 μl): 25 μl of 2×PFU mix, 1.5 μl of each primer V3F / V3R (1 μM), 2 μl of genomic DNA, and 20 μl of sterile water. The PCR reaction conditions were: 95°C pre-denaturation for 5 min; 95°C denaturation for 30 s, 54°C annealing for 30 s, 72°C extension for 3 min, 20 cycles; 72°C final extension for 10 min.
[0046] 2. Obtain detailed sequence information through sequencing, and use NCBI's Blast tool to compare and screen the obtained sequences to obtain highly conserved fragments. The matching rate of Cronobacter is as high as 98%, but that of non-Cronobacter Less than 80% is the standard;
[0047] 3. Select the sequences at both ends, and use the last base ...
Embodiment 2
[0050] Example 2: PCR primer specificity verification
[0051] 1. Primer design:
[0052] Taking the specific sequence obtained by screening in Example 1 as the target sequence as a template, the Cronobacter specific amplification primers were designed, and the primer sequences were as follows:
[0053] VicF1: 5'-TCGTGCTGCGAGTTTGAGAG-3' (SEQ ID NO.2)
[0054] VicR1: 5'-CCTCGCGTGCTCACACAG-3' (SEQ ID NO.3)
[0055] VicF2: 5'-AGAGACTCTGACACACCGCG-3' (SEQ ID NO.4)
[0056] VicR2: 5'-AATGAGTGAAAGGCGTTACCG-3' (SEQ ID NO.5)
[0057] 2. Verification of primer specificity
[0058] Using the genomic DNA of 31 strains of Cronobacter and 21 strains of non-Cronobacteria as templates, the specificity of primers VicF1 / VicR1 and VicF2 / VicR2 was verified by nested PCR amplification detection. The results are shown in Table 1. The steps are as follows:
[0059] Take the genomic DNA template of each strain for the first round of nested PCR amplification, PCR reaction system (25 μl): 2×Taq m...
Embodiment 3
[0069] Example 3: Sample detection application of silica magnetic microspheres
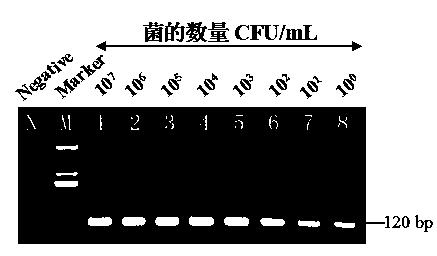
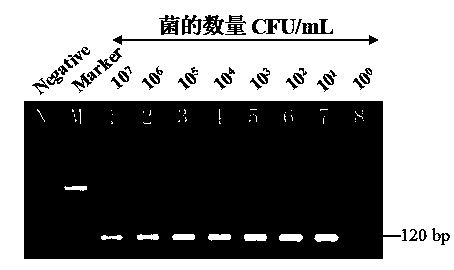
[0070] 1. Detection of pure culture samples of different concentrations of Cronobacter
[0071] Using silica magnetic microspheres (Magpearl Silica, 0.3-0.5 μm, 10 mg / ml, purchased from Zhengzhou Inno Biotechnology Co., Ltd.) and nested PCR technology for specific detection of Cronobacter pure culture samples. The pure culture of Cronobacter was diluted with sterile 0.01M PBS gradient to obtain a concentration gradient of bacteria: 10 7 ,10 6 ,10 5 ,10 4 ,10 3 ,10 2 ,10 1 ,10 0 For the sample of CFU / ml, add 1ml of bacteria dilution solution of different concentrations to each 2ml centrifuge tube in turn as the sample to be tested, and add 1ml of sterile water to the 2ml centrifuge tube as a negative control. Place the centrifuge tube in a boiling water bath for 10 minutes to lyse the cells to release genomic DNA; add 700uL high salt solution (3M NaCl, 2M KCl) and 10uL silica magnetic micro...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com