Method for detecting nucleic acid based on exponential hairpin assembly and colorimetry
A technology for exponential card issuing assembly and card issuing, applied in the field of molecular biology, can solve the problems of difficult grass-roots promotion and application, long amplification reaction time, non-specific amplification, etc., and achieves the effects of high sensitivity, good specificity and short reaction time.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
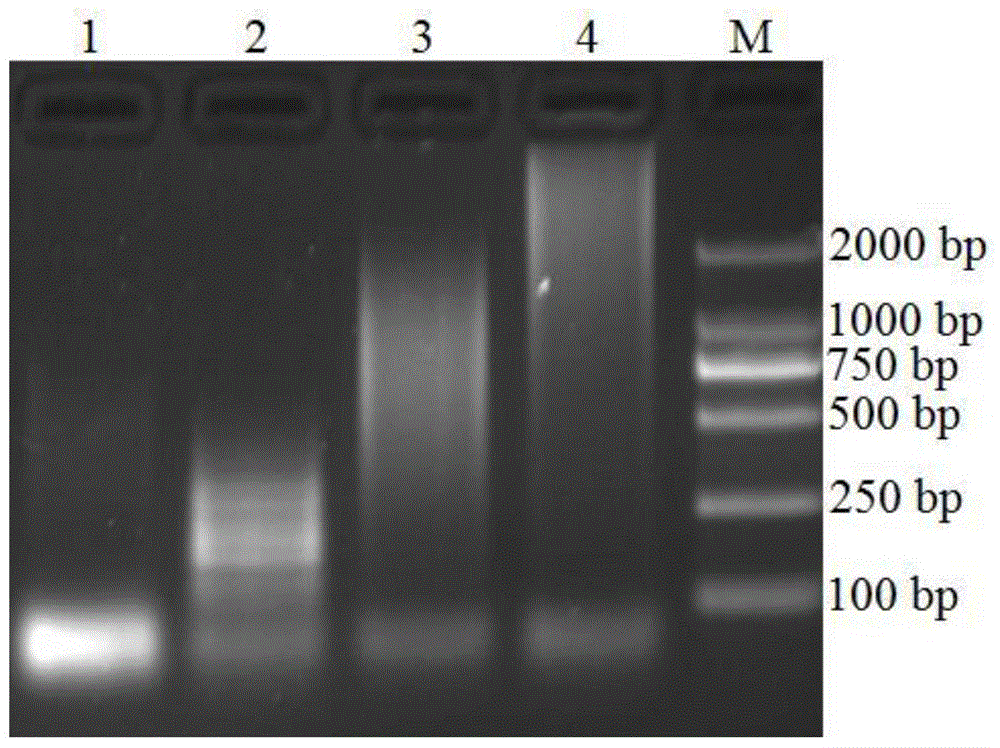
[0035] Embodiment 1: the feasibility of verification method and the correctness of its principle
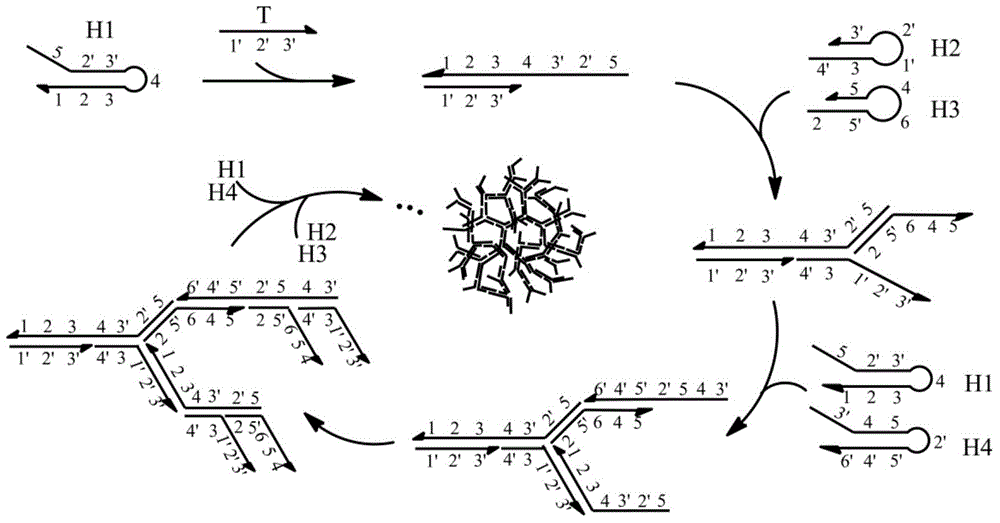
[0036] In this example, the exponential assembly reaction between the target strand and the four hairpin DNAs can form a large DNA polymer in a relatively short period of time, and the speed and molecular weight of the generated product are much higher than that of the linear assembly reaction. Compare the electrophoresis results to verify the method The feasibility and correctness of the principle.
[0037] The principle of index card issuing assembly experiment is as follows: figure 1 As shown, the target nucleic acid molecule hybridizes with the foothold sequence of the first hairpin DNA, and under the drive of free energy, the first hairpin DNA is opened to expose the sequences that can combine with the second and third hairpin DNA ; This sequence hybridizes with the foothold sequences of the second and third hairpin DNAs respectively, and opens the second and third hairpin ...
Embodiment 2
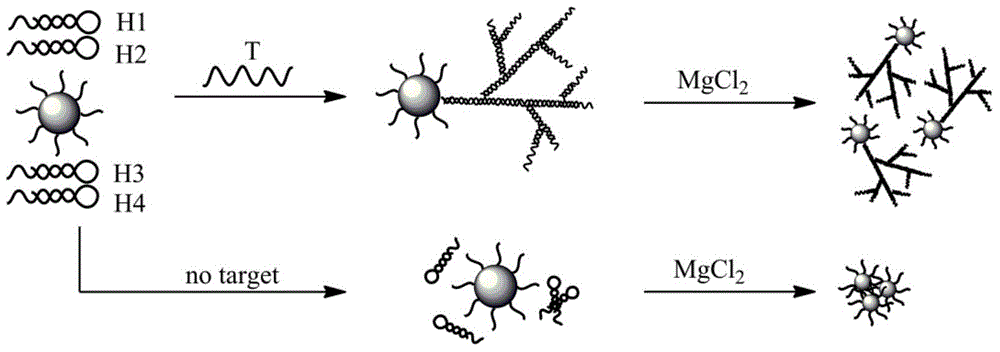
[0047] Example 2: Rapid detection of target DNA using index hairpin assembly colorimetry
[0048] In this example, different concentrations of target DNA were detected to verify the sensitivity of the nucleic acid detection method stated in the present invention.
[0049] The experimental principle of index hairpin assembly colorimetric rapid detection of target DNA is as follows: figure 2 As shown, the EHA reaction can realize the advantage of exponential signal amplification of DNA and the colorimetric properties of nano-gold to achieve rapid and sensitive detection of target DNA. When the target nucleic acid molecule exists, one end of the target nucleic acid molecule can complementarily pair with the sulfhydryl DNA base modified on the gold nanoparticles, and the other end of the target nucleic acid molecule acts as the initiator chain to initiate the EHA reaction, and the DNA can be rapidly synthesized in a short period of time. Growth, so that the surface of nano-gold ...
Embodiment 3
[0058] Example 3: The specific detection of the target DNA is realized by using the method of colorimetric detection of DNA by index hairpin assembly.
[0059] This embodiment detects that one base is mismatched in the target DNA (mismatched target:
[0060] CACAGACCAGAGCAATCCACAACCTAAACCGTTAGAGCCAAC, ie SEQ ID NO.7), delete a base (deleted target: CACAGACCAGAGCAATCCACAACCAAACC GTTAGAGCCAAC, ie SEQ ID NO.8), add a base (inserted target: CACAGACCAGAGCAATCCACAACCAGAAACCGTTAGAGCCAAC, ie SEQ ID NO.9), for investigation The ability of the method of the invention to distinguish base differences when detecting nucleic acids.
[0061] Reaction conditions:
[0062] Take 75 μL of sulfhydryl DNA (SEQ ID NO.6) modified gold nanoparticles, and add 5 μL to the system respectively to a final concentration of 5×10 -10 M's target DNA T (SEQ ID NO.5), 5 x 10 -10 M mismatched target (SEQ ID NO.7), 5×10 -10 M deleted target (SEQ ID NO.8), 5×10 -10 M inserted target (SEQ ID NO.9), followed b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com