Perylene excimer-based detection method for methylase activity and screening method of methylase inhibitor
A technology of methylase activity and methylase, applied in the field of screening of methylase inhibitors, can solve the problems of high cost, time-consuming, low sensitivity, etc. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
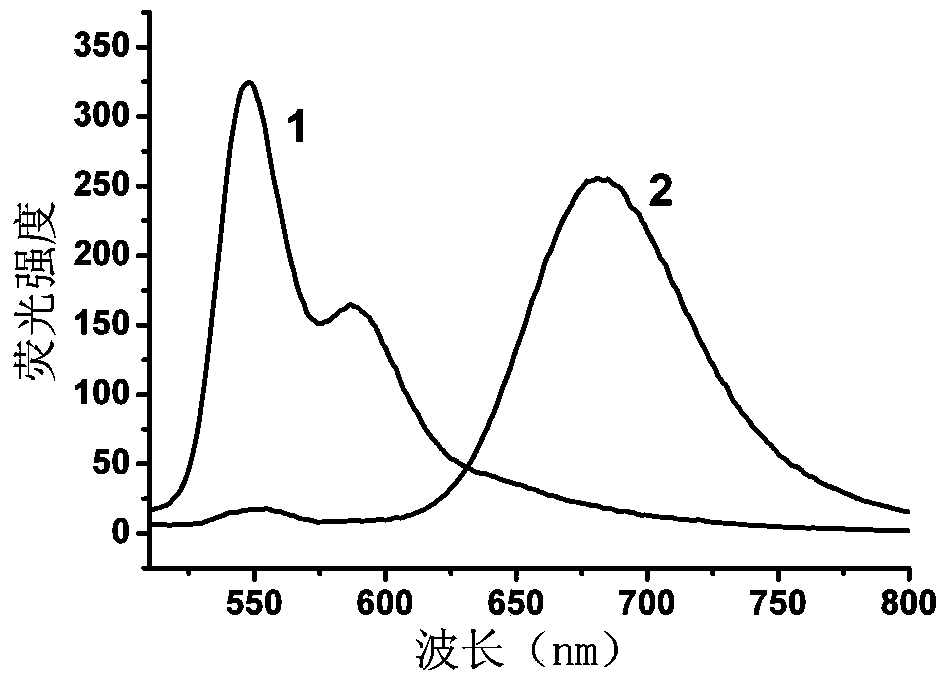
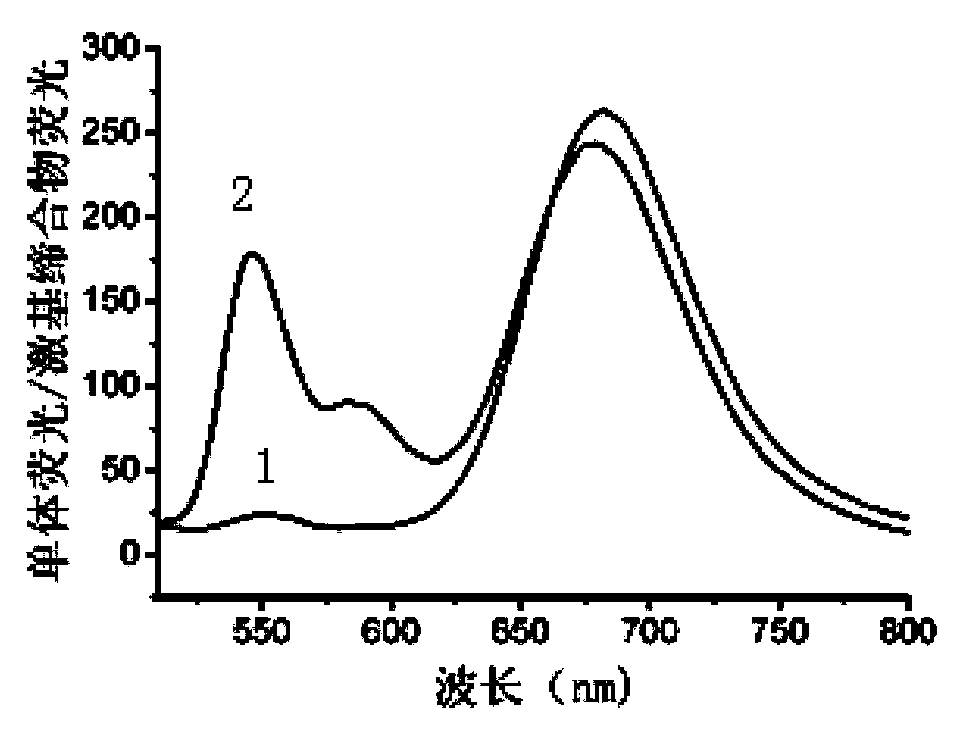
[0066] Mix equal amounts of nucleic acid strands DNA-a (5'-GTTGGGATCGAGAAGddC-3') and DNA-b (5'-CTTCTCGATCCCAACddC-3') in buffer solution [2mMTris-HAc, 5mMKAc, 1mMMg(Ac) 2 ,pH7.9] in a water bath at 90°C for 10 minutes, then cooled to room temperature to obtain double-stranded DNA (dsDNA-1);
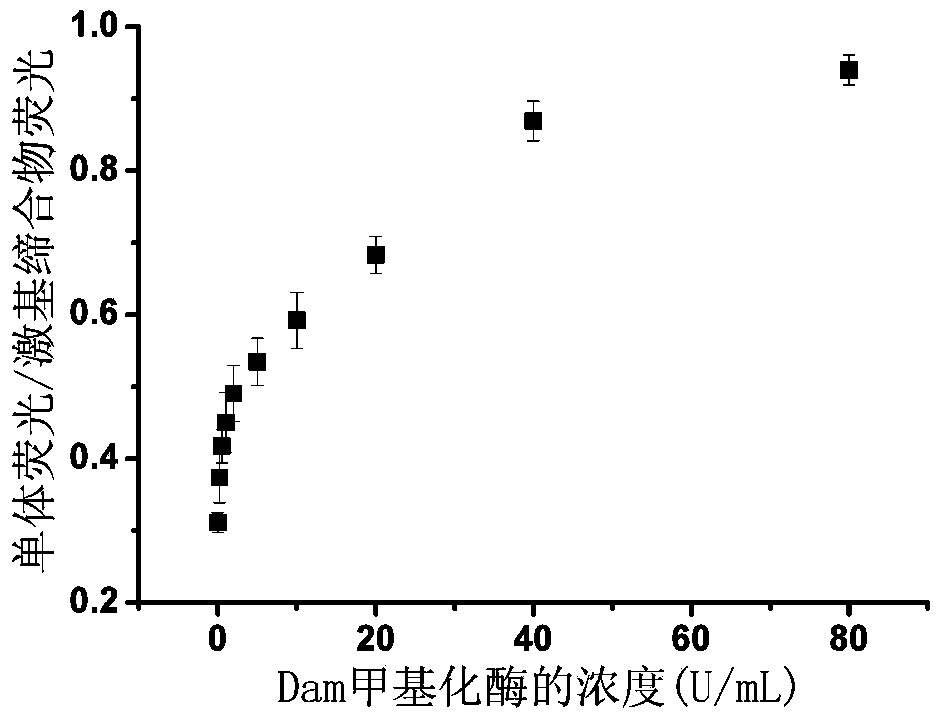
[0067] In a 50μL enzyme reaction system, 400nM dsDNA-1, 2.5μL reaction buffer [10×buffer: 200mM Tris-HAc, 500mMKAc, 100mMMg(Ac) 2,10mMDTT,pH7.9], 160μS-adenosylmethionine (SAM), 200U / mL restriction endonuclease (DpnI) and different concentrations of methylase Dam (concentrations were 0, 0.2, 0.5, 1, 2, 2.5, 10, 20, 40, and 80 U / mL), placed at 37°C for 2 hours, and then placed in a 90°C water bath for 10 minutes to inactivate methylases and restriction enzymes, Finally cooled to room temperature to obtain a mixed solution;
[0068] Add 8 U of terminal deoxynucleotidyl transferase (TdT), 200 μM deoxyribonucleoside triphosphate (dNTP), and 5 μL of TdT reaction buffer [5 μL, 10× buffer: 12...
Embodiment 2
[0077] Mix equal amounts of nucleic acid strands DNA-a (5'-GTTGGCCGGGAGAAGddC-3') and DNA-b (5'-CTTCTCCCGGCCAACddC-3') in buffer solution [2mMTris-HAc, 5mM KAc, 1mMMg(Ac) 2 ,pH7.9] in a water bath at 90°C for 10 minutes, then cooled to room temperature to obtain double-stranded DNA (dsDNA-1);
[0078] In a 50μL enzyme reaction system, 400nM dsDNA-1, 2.5μL reaction buffer [10×buffer: 200mM Tris-HAc, 500mMKAc, 100mMMg(Ac) 2 ,10mMDTT,pH7.9], 160μS-adenosylmethionine (SAM), 200U / mL restriction endonuclease (HpaII) and different concentrations of methylase HpaII (concentrations were 0, 0.2, 0.5, 1, 2, 2.5, 10, 20, 40, and 80 U / mL), placed at 37°C for 2 hours, and then placed in a 90°C water bath for 10 minutes to inactivate methylases and restriction enzymes, Finally cooled to room temperature to obtain a mixed solution;
[0079] Add 8 U of terminal deoxynucleotidyl transferase (TdT), 200 μM deoxyribonucleoside triphosphate (dNTP), and 5 μL of TdT reaction buffer [5 μL, 10× buffe...
Embodiment 3
[0084] Mix equal amounts of nucleic acid strands DNA-a (5'-GTTGGCGGAGAAGddC-3') and DNA-b (5'-CTTCTCCGCCAACddC-3') in buffer solution [2mMTris-HAc, 5mMKAc, 1mMMg(Ac) 2 ,pH7.9] in a water bath at 90°C for 10 minutes, then cooled to room temperature to obtain double-stranded DNA (dsDNA-1);
[0085] In a 50μL enzyme reaction system, 400nM dsDNA-1, 2.5μL reaction buffer [10×buffer: 200mM Tris-HAc, 500mMKAc, 100mMMg(Ac) 2 , 10mMDTT, pH7.9], 160μS-adenosylmethionine (SAM), 200U / mL restriction endonuclease (BstUI) and different concentrations of methylase M.SssI (concentrations were 0, 0.2, 0.5, 1, 2, 2.5, 10, 20, 40 and 80 U / mL), placed at 37°C for 2 hours, and then placed in a 90°C water bath for 10 minutes to deplete methylases and restriction enzymes live, and finally cooled to room temperature to obtain a mixed solution;
[0086] Add 8 U of terminal deoxynucleotidyl transferase (TdT), 200 μM deoxyribonucleoside triphosphate (dNTP), and 5 μL of TdT reaction buffer [5 μL, 10× bu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com