Heparinase II deletion mutant coding gene and protein thereof
A technology for deletion of mutants and encoded genes is applied in the field of heparinase II deletion mutants encoding genes and their proteins, and can solve the problems of difficult purification, low soluble heparinase activity, and low expression level.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
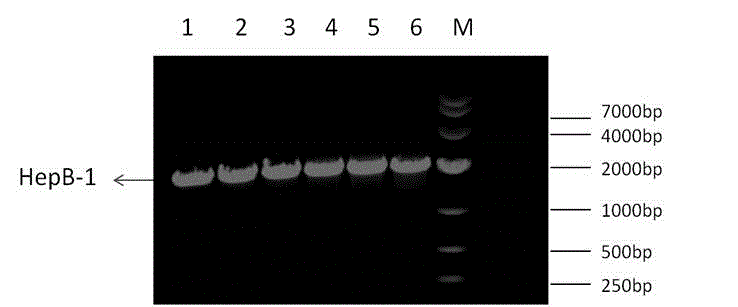
[0024] Example 1. Deletion of the expression of the heparanase II protein HepB-1.
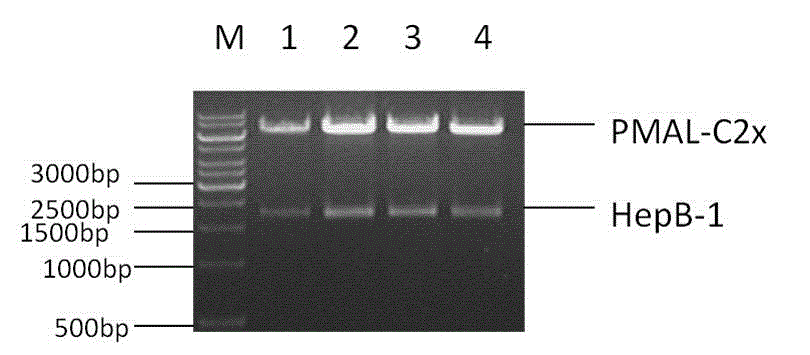
[0025] l. Construction of expression vector pMal-HepB-1.
[0026] For the construction of template PMal-HepB, refer to patent CN 101942024.
[0027] The specific process of constructing the expression vector pMal-HepB-1 is as follows: using the constructed PMAL-HepB as a template, the upstream and downstream primers used are 5' GCCTGAGCTCGCAAACCAAGGCCGATGTG 3 (the bases underlined are the restriction sites of PST I) , 5'-GACTCTGCAGTTAACGATCAACAGTTTCTGAAGTTTTG-3 (bases underlined are SacI restriction sites), respectively introduce PSTI and SacI restriction sites, the amplification reaction system is: 50ng template DNA, 200nmol each primer, 5×PS buffer amplification buffer, 2.5uMol / L of each dNTP, 1 unit of high-retention Pfu enzyme; the amplification program is: denaturation at 95 degrees Celsius for 5 minutes, primer annealing at 50-60 degrees Celsius for 15 seconds, primer extension at 72 deg...
Embodiment 2
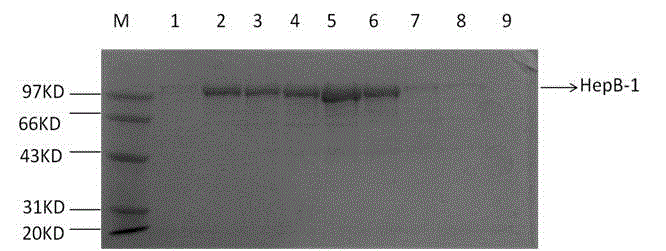
[0031] Example 2. Purification of heparanase I protein HepB-1 by amylose column and determination of heparanase I HepB-1 activity.
[0032] 1. Purification of heparanase Ⅱ protein HepB-1 and heparanase Ⅱ HepB-1.
[0033] The fusion partner (fusion partner) maltose binding protein MBP used in the present invention can achieve one-step separation with amylose affinity adsorption. The specific steps of affinity separation are as follows: centrifuge the bacterial solution at 8000rpm for 10min after induction of 0.1mmol IPTG-induced bacterial cells for 20 hours, discard the filtrate and resuspend in 40ml 20mmol Tris. 5s, intermittent time 5s, ultrasonic 200 cycles), 18000rpm.30min centrifugation. After centrifugation, the supernatant was passed through a 20ml pre-equilibrated amylose affinity separation column at 0.5ml / min, eluted and collected by a gradient of 10mM 0.5ml / min maltose.
[0034] The target protein can be eluted with 10 mM maltose at 10 column volumes. The result i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com