Method for synthetizing D-phenyllactic acid through recombinant Escherichia coli
A technology for recombining Escherichia coli and amino acids, applied in the field of genetic engineering, can solve the problems such as limited production of phenyllactic acid, and achieve the effects of improving affinity, catalytic efficiency, and yield.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
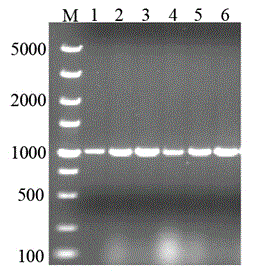
[0041] This example illustrates the construction of pET-28a-ldh.
[0042] Pick a single colony of Lactobacillus plantarum in 10 ml of liquid medium for overnight culture at 30°C and 200 rpm (about 10 h), inoculate the seed culture solution in 10 ml of fresh liquid medium at 1% and cultivate until the late logarithmic period, use Genomic DNA Extraction Kit (Shanghai Sangong Bacteria Genomic DNA Rapid Extraction Kit) was used to extract the genome. Liquid medium (g / L): casein peptone 10, beef extract 10, yeast extract 5, glucose 5, sodium acetate 5, diamine citrate 2, Tween 80 1, KHPO4 2, MgHPO47H2O 0.2, MnSO4H2O 0.05, CaCO3 20, pH 6.8, make up to 1L with distilled water.
[0043] The primer sequences used to construct the expression vector are as follows:
[0044] Upstream primer (including BamHI restriction site): CGC GGATCC ATGAAAATTATTGCATATGC
[0045] Downstream primer (with HindIII restriction site): CCC AAGCTT TTAATCAAACTTAACTTGTG
[0046] Lactate dehydrog...
Embodiment 2
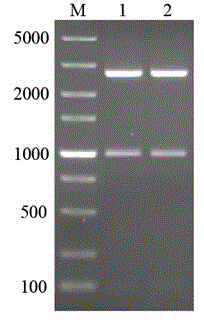
[0051] This example illustrates the construction of pET-28a-ldhY52V.
[0052] Using the extracted and constructed recombinant plasmid pET-28a-ldh as a template and P1 / P2 as primers, PCR was carried out according to the instructions of the site-directed mutagenesis kit, and the reaction product was transformed into competent E. coli JM109, and positive clones were identified by colony PCR and plasmid double-enzyme electrophoresis , and then extract the successfully constructed recombinant plasmid pET-28a-ldhY52A and send it to Sangon for sequencing. After being accurate, it will transform and express the host BL21(DE3) competent, screen positive clones, and express the recombinant strain BL21(DE3). The construction of pET-28a-ldhY52A is completed .
[0053] Using the extracted and constructed recombinant plasmid pET-28a-ldh as a template and P3 / P4 as primers, PCR was performed according to the instructions of the site-directed mutagenesis kit, and the reaction product was trans...
Embodiment 3
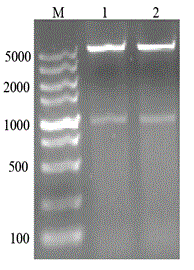
[0056] This example illustrates the transformation of whole cells of recombinant Escherichia coli to synthesize phenyllactic acid.
[0057] Pick a single colony of the recombinant bacteria and inoculate it in LB liquid medium containing 50 mg / L kanamycin sulfate, culture overnight (about 9 h) at 37°C and 200 r / min, and inoculate 1% of the culture medium containing the same concentration of cardamycin In the fresh LB medium of namycin, culture at 37°C and 200 r / min until the OD600 is about 0.6-0.8, add IPTG concentration to 0.6 mmol / L, lower the induction temperature to 25°C, and induce at 200 r / min After 5 h, the bacteria were collected by centrifugation, washed twice with pre-cooled pH 7.0 phosphate buffer, and the prepared sludge was used for whole cell transformation.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com