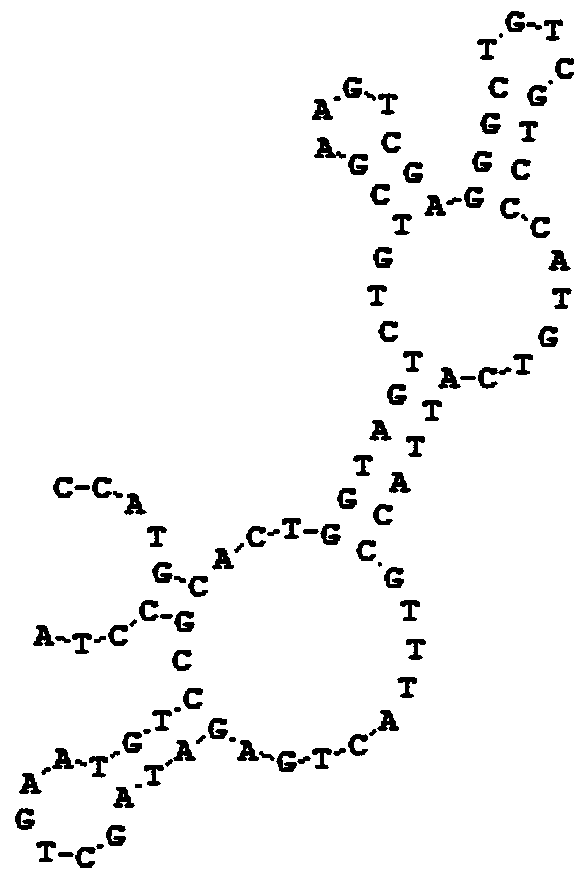Nucleic acid aptamer for combining cholera enterotoxin subunit B and application thereof
A nucleic acid aptamer, enterotoxin technology, applied in the determination/inspection of microorganisms, resistance to vector-borne diseases, DNA/RNA fragments, etc. Obtain the effect of simple, fast, low price and good application prospect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0015] Example 1 Screening of a single-stranded DNA aptamer capable of binding to cholera enterotoxin B subunit
[0016] Using SELEX technology, proceed as follows:
[0017] 1. Synthesis of ssDNA random library and primers
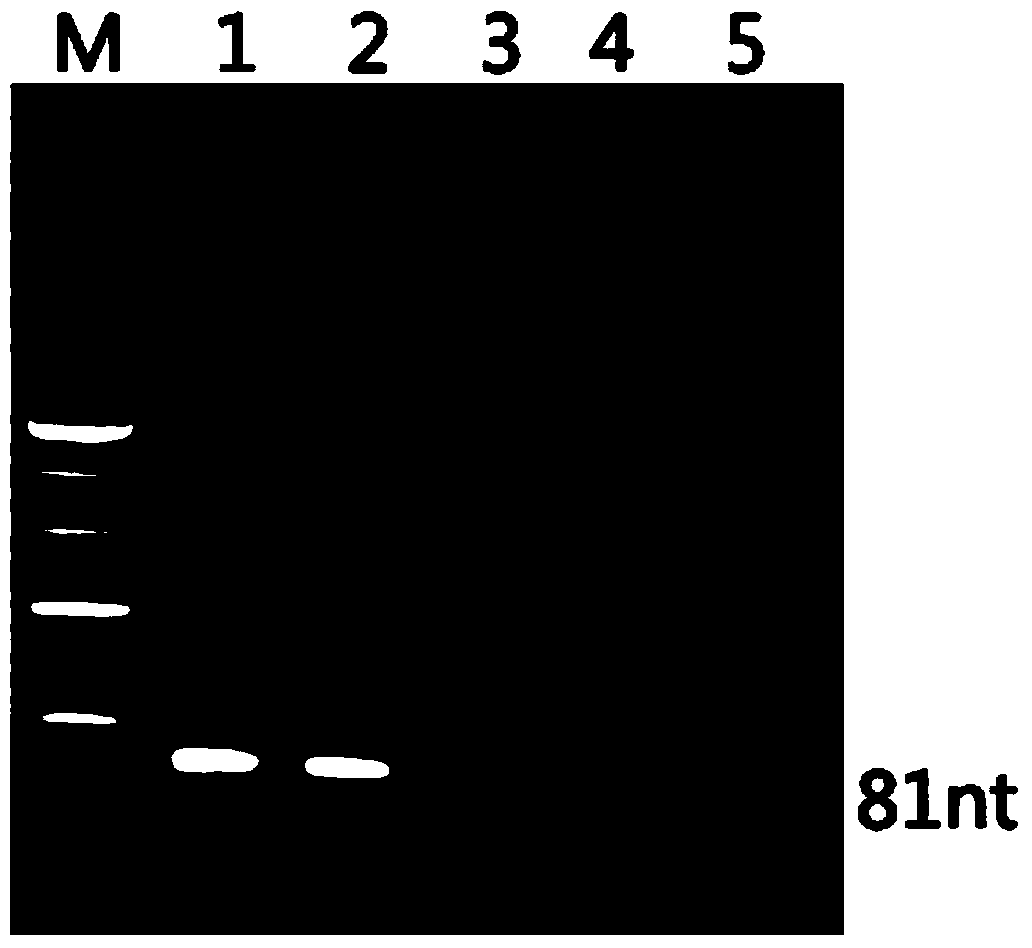
[0018] The total length of the ssDNA library used for SELEX screening is 81nt, in which both ends are fixed sequences bound by primers, and the middle 35 nucleotides are random sequences.
[0019] The library sequences are:
[0020] 5'-CCATGCACTGGTAGTCTGTCGAAGT-(N35)-AGATAGCTGAATGTCCGCCTA-3'.
[0021] The primer sequences for the amplified library were:
[0022] Upstream primer: 5'-CCATGCACTGGTAGTCTGTCGAAGT-3';
[0023] Downstream primer: 5'-TAGGCGGACATTCAGCTATCT-3';
[0024] Biotin-labeled upstream primer: 5’-Biotin-CCATGCACTGGTAGTCTGTCGAAGT-3’
[0025] The random ssDNA library and primers were synthesized by Bao Biological Engineering (Dalian) Co., Ltd. and purified by PAGE.
[0026] 2. SELEX screening process of cholera enterotoxin B subunit apta...
Embodiment 2
[0036] The ssDNA library of embodiment 2 screening and the mensuration of target protein binding rate
[0037] (1) Asymmetric PCR amplification was performed using biotin-labeled upstream primers and ordinary downstream primers to obtain a biotin-labeled ssDNA library at the 5' end. After electrophoresis, the gel was cut and purified, and the concentration was adjusted to 1.5 pmol / μl.
[0038] (2) Coating a 96-well ELISA plate with cholera enterotoxin B subunit protein, 200 ng per well, and setting blank control wells in parallel.
[0039] (3) After high-temperature denaturation at 95°C for 5 minutes, add the labeled ssDNA library of each round to the reaction well for binding reaction with the toxin protein, incubate at 37°C for 2 hours, and discard the liquid in the well.
[0040] (4) wash 3 times with washing buffer (PBS+0.05% Tween20, PBS-T, pH7.4), each 3min, then add freshly prepared streptavidin-horseradish peroxidase conjugate ( HRP-labeled Streptavidin, diluted 1:100...
Embodiment 3
[0044] Example 3 Detection of cholera enterotoxin B subunit by membrane hybridization method using biotin-modified nucleic acid aptamer
[0045] The affinity between the aptamer and cholera enterotoxin B subunit was determined by membrane hybridization. First, re-synthesize the aptamer described in SEQ ID No.1 and label it with biotin at the 5' end, dissolve it with coating buffer (0.05mol / L NaHCO3, pH9.6) and make it 1μM; cut 1 sheet 1.6×3.8cm nitrocellulose membrane strips, drawn into 6 grids with seal oil; numbered 1-6, of which grid 1 is the negative control of BSA (0.5 μg), grid 2 is the blank control (without cholera enterotoxin protein ); the sample volumes of No. 3, 4, 5, and 6 cholera enterotoxin B subunits were: 0.05, 0.1, 0.2, and 0.5 μg, and the sample volume was 1.5 μL. Place the membrane strips in a small square box, add 2ml blocking solution (10% skimmed milk powder) and carry out blocking treatment at room temperature for 2 hours; then wash 2 times with PBS, t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com