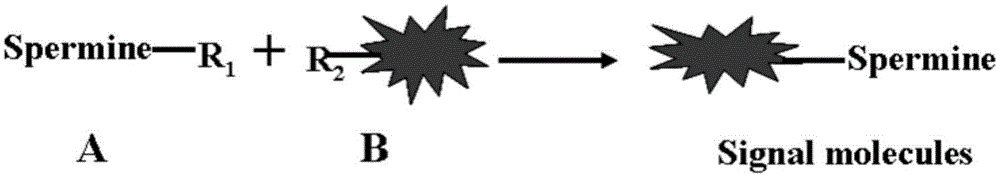
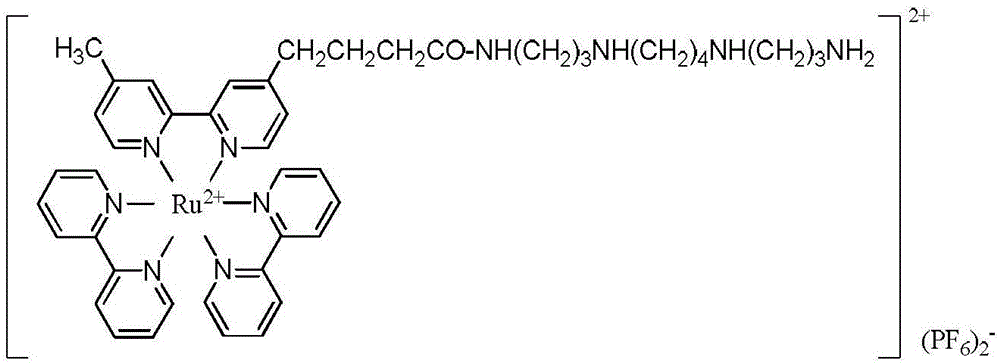Signal labeled molecule for DNA oxidative damage product 8-hydroxydeoxyguanosine and labeling method
A technology of hydroxydeoxyguanosine and signal labeling, which is applied in biological testing, material analysis, material analysis through optical means, etc. It can solve the problems of higher detection value than the real value, difficulty in antibody preparation, and cross-reaction. High labeling rate, good specificity, and easy preparation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Example 1, Synthesis of Signal Marker Molecules
[0042] 1. Take a 50ml two-necked round bottom flask, one of which is protected by a nitrogen balloon, and add 1.28g of 4-methyl-4'-(3-hydroxypropyl)-2,2'-bipyridine, DCC1. 545g, NHS0.575g, DMF15ml, stirred at room temperature for 5h.
[0043] 2. Take another 50ml two-necked round bottom flask, one of which is protected by nitrogen gas, and the other is equipped with a 50ml constant pressure dropping funnel, and 10.1g of spermine (diaminopropyltetramethylenediamine) and 10ml of DMF are added to the bottle. Transfer the reaction solution obtained after the reaction in Step 1 into a constant pressure dropping funnel under nitrogen protection, and slowly add it dropwise to the reaction flask under vigorous stirring. After the dropwise addition, continue to react for 1 h, remove DMF under reduced pressure, and add Add 2M hydrochloric acid and dichloromethane to the compound, separate liquid, wash twice with saturated sodium ...
Embodiment 2
[0045] Embodiment 2, gold electrode surface DNA incubation time optimization
[0046] The DNA modified with sulfhydryl groups at the end was dissolved in the buffer containing fresh TCEP-Tris, 4 μl was dropped onto the surface of the treated gold electrode, covered with the electrode cap, and incubated for 8h, 16h and 40h, respectively. After the end, wash with deionized water, blow dry with nitrogen, soak in 200ul of mercaptohexanol and incubate for a period of time, wash with deionized nitrogen and dry with nitrogen. First place the modified electrode in a Tris solution containing 3.5 μM RuHex to scan CV at a scan rate of 50 mV / s. Then washed with water and placed in a Tris solution containing 50 μM RuHex to scan CV at a scan rate of 50 mV / s. Comparing the CV diagrams under different incubation times, it was found that the DNA assembly on the electrode surface was saturated and relatively stable after 40 hours of incubation.
Embodiment 3
[0047] Embodiment 3, gold electrode surface DNA concentration optimization
[0048] Five different concentrations of 0.1, 0.5, 1, 2, and 5 μM DNA solutions with end-modified sulfhydryl groups were selected and added dropwise to the surface of the gold electrode, and incubated at room temperature. Wash with deionized water, dry with nitrogen, soak in mercaptohexanol for a period of time, wash with deionized water and dry with nitrogen. The modified electrode was placed in a Tris solution containing 50 μM RuHex to scan CV at a scan rate of 50 mV / s. Comparing the CV plots of different concentrations of DNA, it was found that the concentration of DNA reached saturation when the concentration was 2 μM.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com