Method for screening beta-glucosaccharase gene from mildewed sugarcane leaves based on metagenomic technology
A technology of glucosidase and screening method, which is applied in the field of beta-glucosidase gene and recombinant beta-glucosidase, and can solve the problems of sensitivity and low activity of hydrolyzed products.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
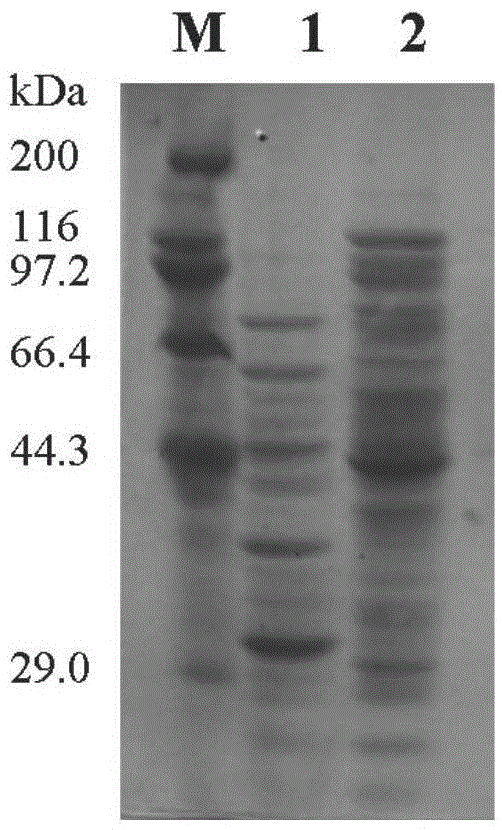
[0042] Example 1 Establishment of metagenomic library and acquisition of positive clones, gene cloning and expression
[0043] 1) Extraction of microbial metagenomic DNA from moldy sugarcane leaves: Collect 10 g of moldy sugarcane leaves, cut them to a length of 1-3 cm, and put them into a 250 ml Erlenmeyer flask. Add enrichment medium (g / L: sodium carboxymethylcellulose, 5; NaNO 3 ,2;K 2 PO 3 ,1; KCl,0.5; MgSO 4 ,0.5; FeSO 4 , 0.01.) 100ml, cultured with shaking at 30°C for 72h. The bacteria were collected by centrifugation, and the bacteria were resuspended in 1.35ml DNA extraction buffer (100mM Tris-HCl[pH8.0], 100mM EDTA-Na 2 [pH8.0], 100mM phosphate buffer [pH8.0], 1.5MNaCl, 1% CTAB), mix well, add 0.15ml 20% SDS, bathe in 65℃ water for 2 hours, during this period, turn it upside down 10 times every 20min and freeze it in liquid nitrogen 2min; centrifuge at 6000g for 10min, collect the supernatant, add equal v of chloroform, mix up and down; centrifuge at 6000g for ...
Embodiment 2
[0065] The study of the enzymatic properties of embodiment 2 recombinant β-glucosidase
[0066] Take 100μl crude enzyme solution, add 100μlpNPG, bathe in water at 35℃ for 15min, add 100μl0.5M Na2CO 3 , while using the inactivated crude enzyme solution as a control, measure the light absorbance at 405nm.
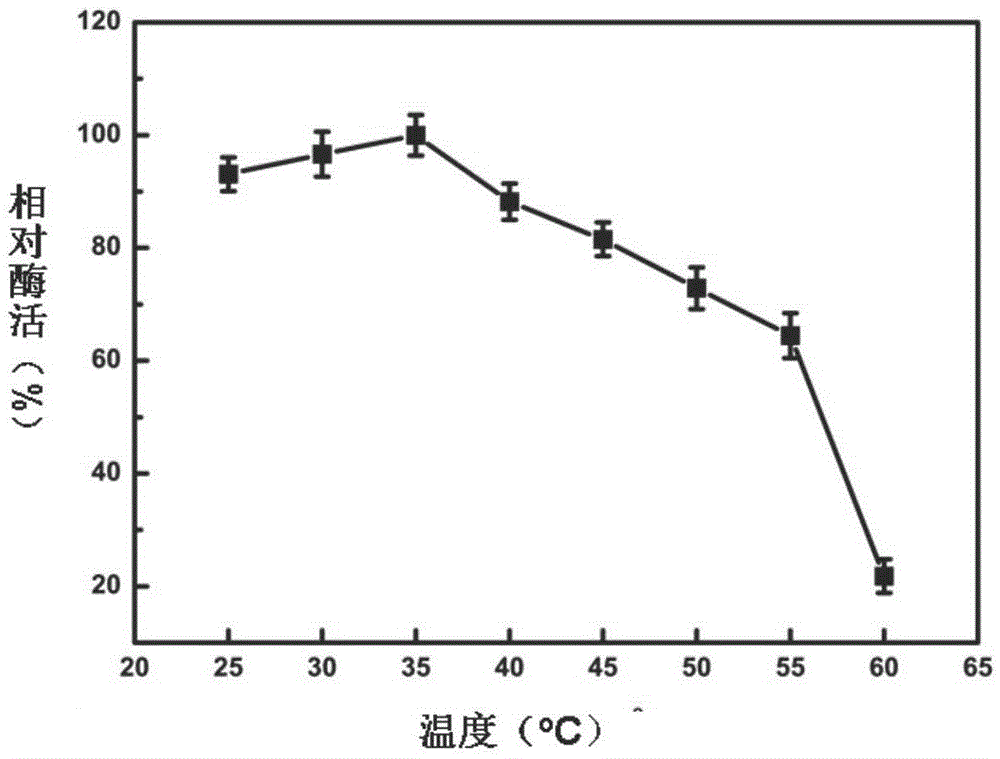
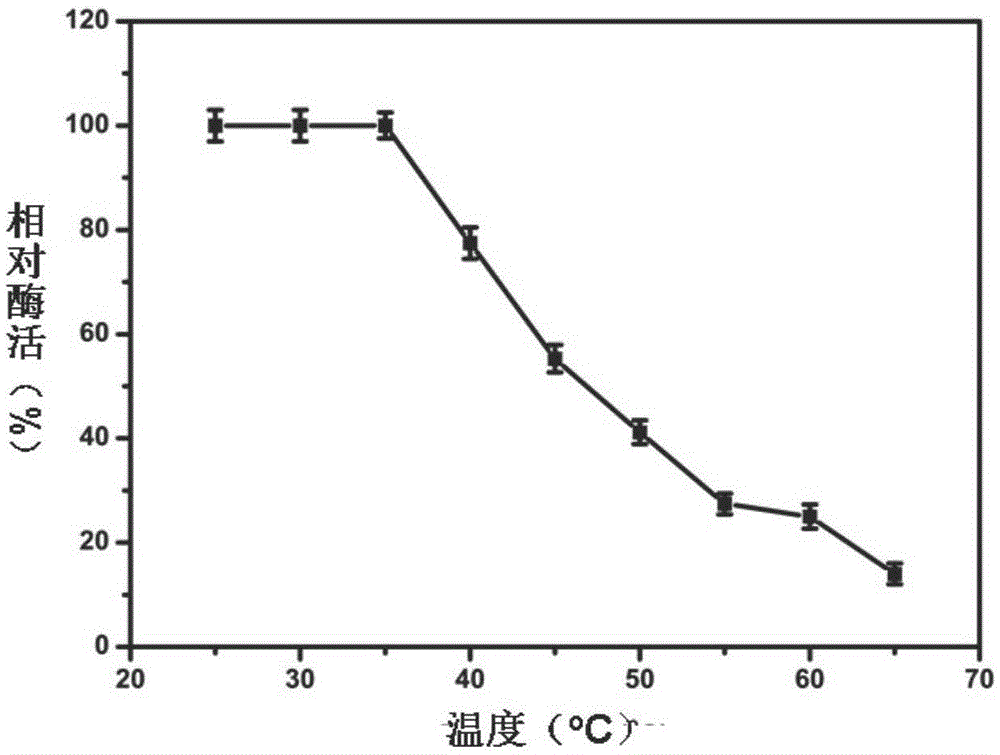
[0067] 1) Optimum reaction temperature and temperature stability of recombinant β-glucosidase Glu417
[0068] Under the condition of reaction temperature of 20-60° C., the enzyme activity was measured according to the above method, and the optimum reaction temperature was obtained (recorded as 100% when the enzyme activity was the highest). Incubate at 25, 30, 35, 40, 45, 50, 55, 60 and 65°C for 2 hours, measure the remaining enzyme activity at 35°C, and obtain the thermal stability of the enzyme. The result is as figure 2 with 3 As shown, the optimum reaction temperature of Glu417 is 35°C. When the temperature is lower than 35°C, Glu417 is stable. As the temperature rises...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com