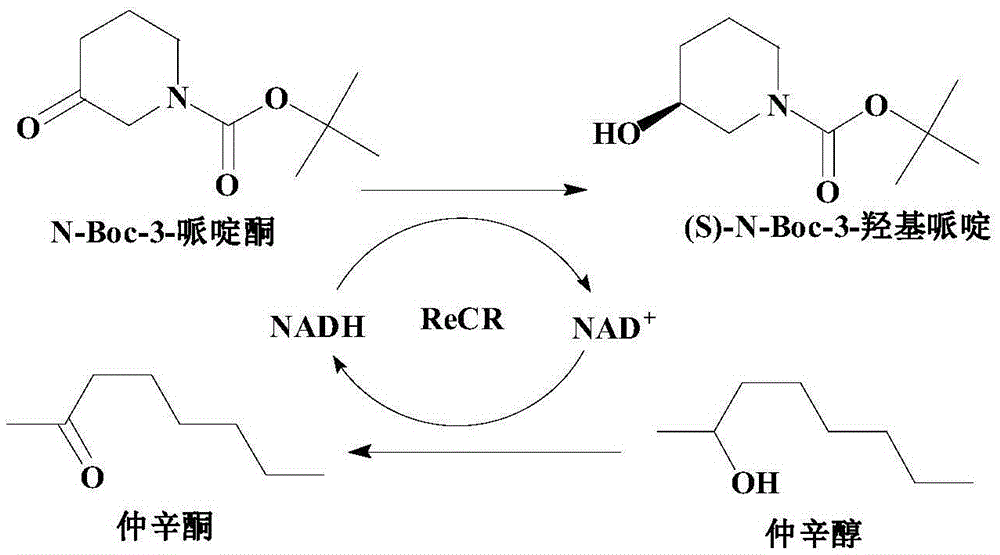
Recombinant carbonyl reductase ReCR, encoding gene, vector, engineering bacterium and application thereof
A technology that encodes genes and reductases, applied in genetic engineering, oxidoreductases, applications, etc., can solve problems such as reducing catalytic efficiency, and achieve the effects of efficient circulation, high optical purity, and specific stereoselectivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1: Construction and induced expression of recombinant carbonyl reductase ReCR Escherichia coli genetically engineered bacteria
[0034] (1) Recombinant carbonyl reductase ReCR Escherichia coli genetic engineering bacteria
[0035] Using the genomic DNA of Rhodococcus erythropolis WZ010 strain as a template, the designed primers F1 and R1 were used for PCR amplification. The amplification system is shown in Table 1.
[0036] Table 1 PCR amplification reaction system
[0037]
[0038] The primers are as follows: F1, 5'-ATGAAGGCAATCCAGTACAC-3'; R1, 5'-CTACAGACCAGGGACCACA-3'. The PCR reaction process is as follows: pre-denaturation at 94°C for 5 minutes; after that, denaturation at 94°C for 30 s, renaturation at 55°C for 30 s, and 72°C for 1 min as a cycle, repeating this cycle 30 times; finally, 72°C for 10 min. PCR products were detected by 0.8% agarose gel electrophoresis. figure 2 A bright band can be seen at about 1000bp, which is consistent with the the...
Embodiment 2
[0045] Embodiment 2: the activity measurement of recombinant carbonyl reductase ReCR and its genetically engineered bacteria
[0046] Enzyme activity was determined by spectrophotometry, and the change in absorbance at 340nm was measured at the optimum temperature and pH, and repeated twice; the optimum temperature and pH for alcohol oxidation were 50°C and 10.0, respectively, while the optimum for ketone reduction was 50°C and 10.0, respectively. The temperature and pH were 60°C and 6.0, respectively. Standard reducing activity assay system (2.5 mL): 50 mMPIPES buffer (pH 6.0), 10 mM N-Boc-3-piperidone, 0.4 mM NADH, recombinant carbonyl reductase ReCR enzyme solution prepared by the method in Example 1 2 μg / mL. Standard oxidation activity assay system (2.5mL): 50mM CAPSO buffer (pH10.0), 50mM 2-octanol, 0.4mM NAD + , the recombinant carbonyl reductase ReCR enzyme solution prepared by the method in Example 1 was 2 μg / mL. Enzyme activity unit definition: at the optimum reacti...
Embodiment 3
[0048] Embodiment 3: Optimum, pH optimum temperature and thermostability of recombinant carbonyl reductase ReCR
[0049] (1) adopting the common buffer system configuration buffer solution of 50mM in the concentration of different pH buffer ranges: MES buffer solution, pH=5.5,6.0; PIPES buffer solution, pH=6.1,6.5,7.0,7.5; Tris-HCl buffer solution, pH=7.5, 8.0, 8.5, 9.0; CAPSO buffer, pH=9.0, 9.5, 10.0; CAPS buffer, pH=10.0, 10.5, 11.
[0050] Reducing activity assay system (2.5 mL): 50 mM buffer, 10 mM N-Boc-3-piperidone, 0.4 mM NADH, recombinant carbonyl reductase ReCR enzyme solution prepared by the method in Example 1 2 μg / mL. Oxidation activity assay system (2.5mL): 50mM buffer, 50mM 2-octanol, 0.4mM NAD + , the recombinant carbonyl reductase ReCR enzyme solution prepared by the method in Example 1 was 2 μg / mL. The reaction temperature for alcohol oxidation is 50°C, while the reaction temperature for ketone reduction is 60°C. The activities of the recombinant carbonyl ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com