Oxalate decarboxylase and recombinant expression method of oxalate decarboxylase
A technology of oxalate decarboxylase and expression method, which is applied in the direction of recombinant DNA technology, biochemical equipment and methods, enzymes, etc., can solve the problems of OXDC incapability, recombinant expression of Escherichia coli, and reduce production costs, so as to improve expression efficiency and strong resistance Pepsin degradation function, effect of reducing production cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Embodiment 1: Screening and activity range test of oxalate decarboxylase
[0034] The inventor has screened more than a hundred species, including Agrocybeaegirit, Tricholoma Lobayensc Heim, Agrocybe Cylindracea, Bacillus subtilis, Coriolus versicolor, brown rot fungus ( Postiaplacenta), cyanobacteria (Cyanobacteria) and other species screened a series of oxalate decarboxylases with high activity in the gastrointestinal pH environment (pH1.5-7.5).
[0035] The screening method is as follows:
[0036] Inoculate and shake the flask to culture the cells, adjust the pH to 2.5-3.0 with phosphoric acid to induce the expression of OXDC, collect the cells, weigh 2g each of the wet cells, add PBS with pH 3.0 and mix well, then crush with a high-speed homogenizer at 9500rpm 18s, add centrifuged bacteria or homogenate supernatant to oxalic acid reaction solution (pH1.5~7.5) of different pH, react at 37℃, 1000rpm for 30min; 100μL 2.5mol / LH 2 SO 4 Stop the reaction. Centrifuge a...
Embodiment 2
[0042] Example 2: Cloning and recombinant expression of oxalate decarboxylase gene
[0043] The oxalate decarboxylase-producing species with excellent performance screened above were cultured in shake flasks and induced for enzyme production. The bacteria were collected at the mid-stage of enzyme production to extract their total RNA and reverse-transcribed into cDNA. Merger primers were designed, and RACE was used to (RapidAmplificationofcDNAEnds) PCR carries out cDNA amplification, and the operation is strictly in accordance with the SMARTer of Clontech company TM The RACE cDNA Amplification Kit operation manual was implemented, and the 5' and 3' universal primers and gene localization primers (OXDCGSPprimer) used in it are shown in Table 2. The amplified gene was sequenced and screened to obtain the gene sequence, and the translated protein sequence is shown in Table 3.
[0044] Table 2. Degenerate primer sequences
[0045]
[0046] Low pH stable oxalate decarboxylase ...
Embodiment 3
[0048] Example 3: Escherichia coli recombinant expression of OXDC
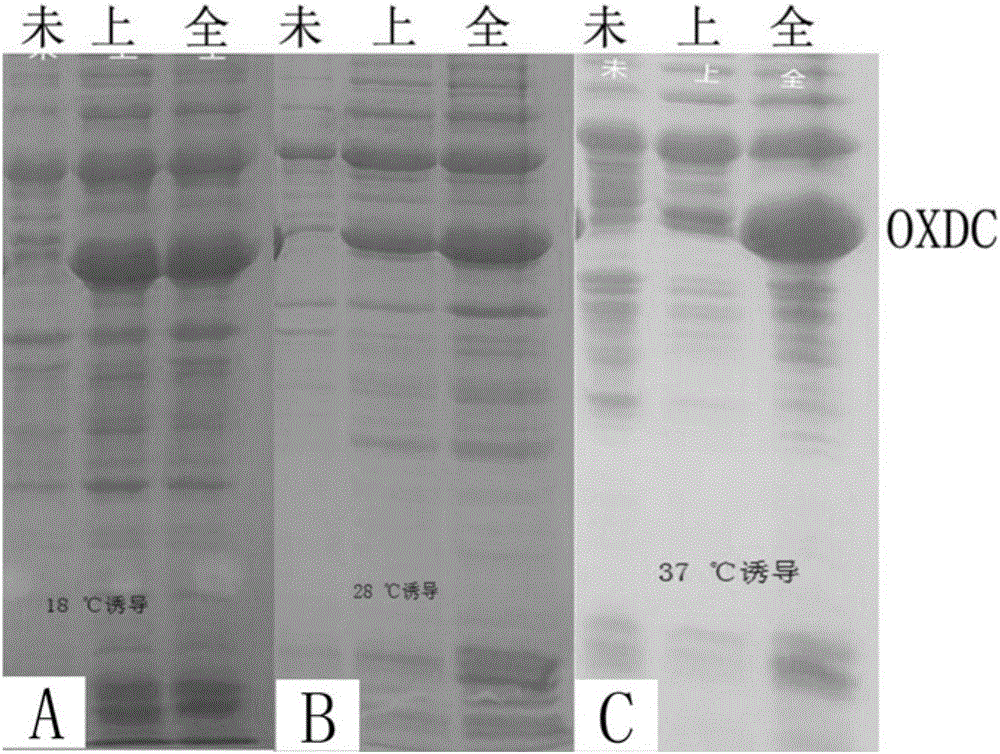
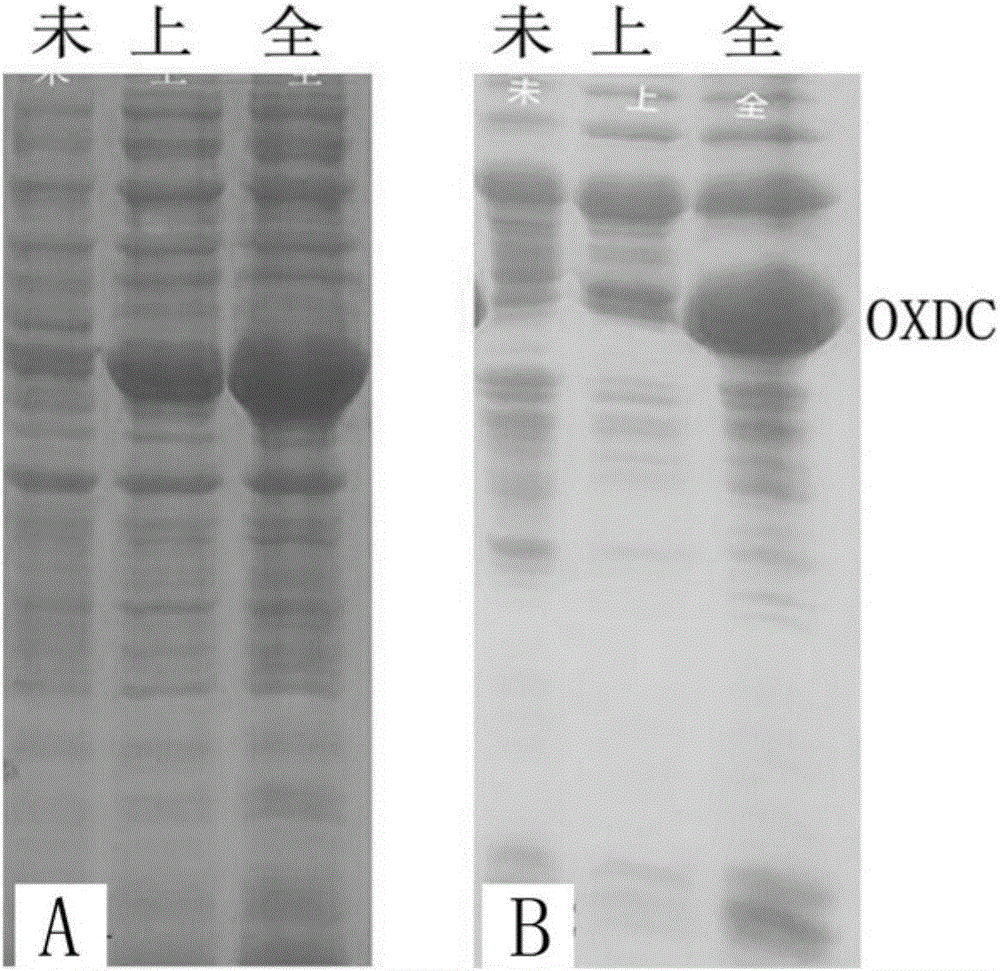
[0049] The present inventors carried out recombinant expression in E. coli system on the low pH stable OXDC (including the previously reported Flammulina velutipes OXDC) cloned by screening. The OXDC gene of Flammulina velutipes has been reported for more than 10 years, see literature (MeenuKesarwani, et.alOxalateDecarboxylase from Collybiavelutipes, THE JOURNALOF BIOLOGICALCHEMISTRY, 2000), the OXDC gene of Flammulina velutipes has not been successfully recombinantly expressed in the E. coli expression system so far. In view of this, this example takes Flammulina velutipes OXDC as an example to illustrate the recombinant expression method of OXDC in Escherichia coli.
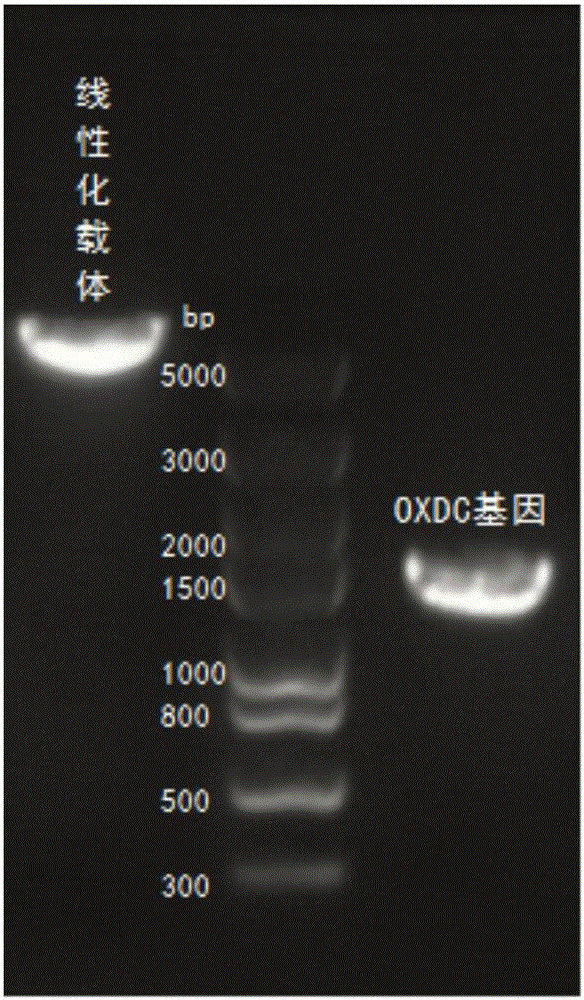
[0051] Design cloning primers (table 4) according to expression vector and Flammulina velutipes OXDC gene sequence, need the coding sequence of signal peptide part to be removed during primer design, directly amplify ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com