Expression vector and expression method of calf intestinal alkaline phosphatase
An expression vector and expression method technology are applied in the field of expression vector of calf intestinal alkaline phosphatase to achieve the effects of accurate expression and good sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] The construction of embodiment 1 expression vector
[0044] 1) Artificially synthesize the nucleotide sequence shown in SEQ ID NO: 2, and digest it with NdelI-HindIII enzyme, and the digestion condition is 37 degrees. The digested product was subjected to agarose gel electrophoresis, and a nucleotide fragment with a size of 1545 bp was recovered from the gel.
[0045] 2) The PMAL-c5x vector is digested with NdelI-HindIII enzyme, and the digestion condition is 37 degrees. The digested products were subjected to agarose gel electrophoresis, and fragments with a size of about 1500 bp were recovered from the gel.
[0046] 3) Ligate the digested linear vector and the target fragment with T4 DNA ligase at a ligation temperature of 16°C overnight.
[0047] 4) The ligation product was transformed into Escherichia coli DH5α, cultured in LB medium containing ampicillin antibiotic, and positive colonies were picked for colony PCR identification. Propagate the colonies with posi...
Embodiment 2
[0073] Embodiment 2 fusion protein expression
[0074] 1. The strains constructed in Example 1 and Comparative Examples 1 to 4 were induced to express the target protein, and detected, the method was as follows:
[0075] 1), Shake the strain in 3ml LB medium to OD 600 0.6, add IPTG, IPTG concentration is 0.3mmol / L, 37°C, 140rpm induction overnight;
[0076] 2) SDS-PAGE electrophoresis to detect the expression of the target protein of each strain, and to detect inclusion bodies.
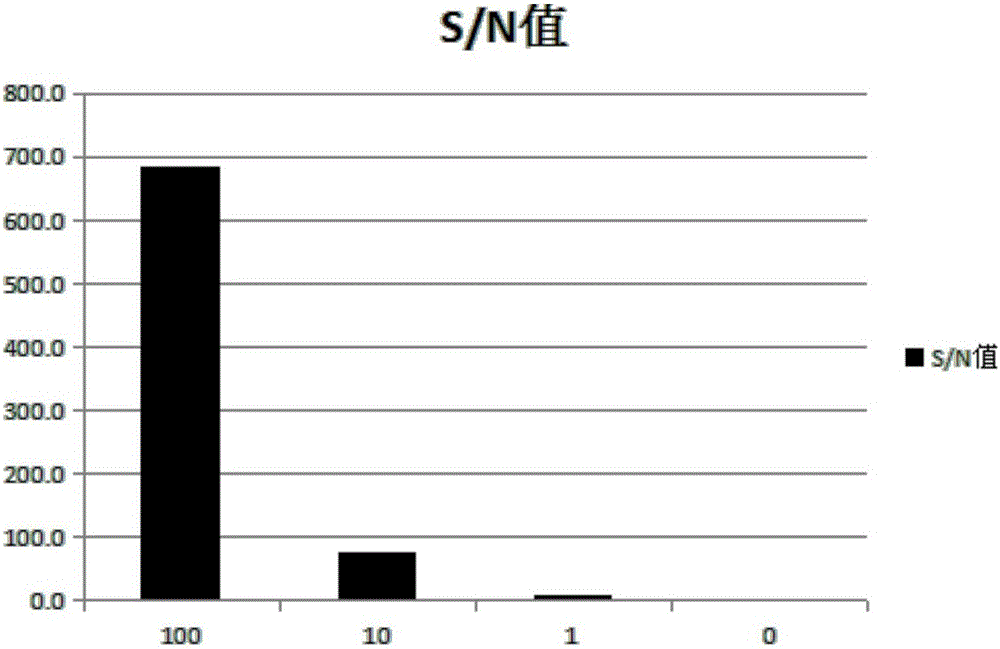
[0077] The results show that: after the strains of Example 1 and Comparative Examples 1 to 2, the fusion protein was successfully expressed in the supernatant formed after the broken bacteria; while Comparative Example 3 and Comparative Example 4 also expressed the fusion protein, However, its protein formed inclusion bodies and did not appear in the supernatant.
[0078] 2, the bacterial classification of embodiment 1, comparative example 1~2 is carried out expansion culture, and detects, and meth...
Embodiment 3
[0088] Preparation and application of embodiment 3 probes
[0089] Taking the CIAP protein extracted and obtained in the prior art as a control, and taking the CIAP fusion protein expressed by the strain obtained in Example 1 as the experimental object, the two were respectively coupled with a probe whose sequence is AGGTCTCTGCTTAGAGGTACTT, and the probe is used for SAT Technical Report Detection Probes. For coupling methods, please refer to patents 6686461, 6800728, 7102024, 7173125, and 7462689 with product instructions attached.
[0090] Implementation process:
[0091] The application of probe labels in SAT technology;
[0092] Step 1 designs a nucleic acid sequence of a coating primer (5'-NH2-GAGCGGATATATGCTTAGGAAT-3'), synthesizes the sequence and dissolves it in a TE buffer solution with a pH of 8.0, and dilutes the primer to 3-5ug / ml;
[0093] Step 2 Prepare the coating buffer, according to the components in the formula table, sodium dihydrogen phosphate, disodium h...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com