A kind of β-glucosidase gene and its application
A technology of glucosidase and gene, applied in the field of microorganisms
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
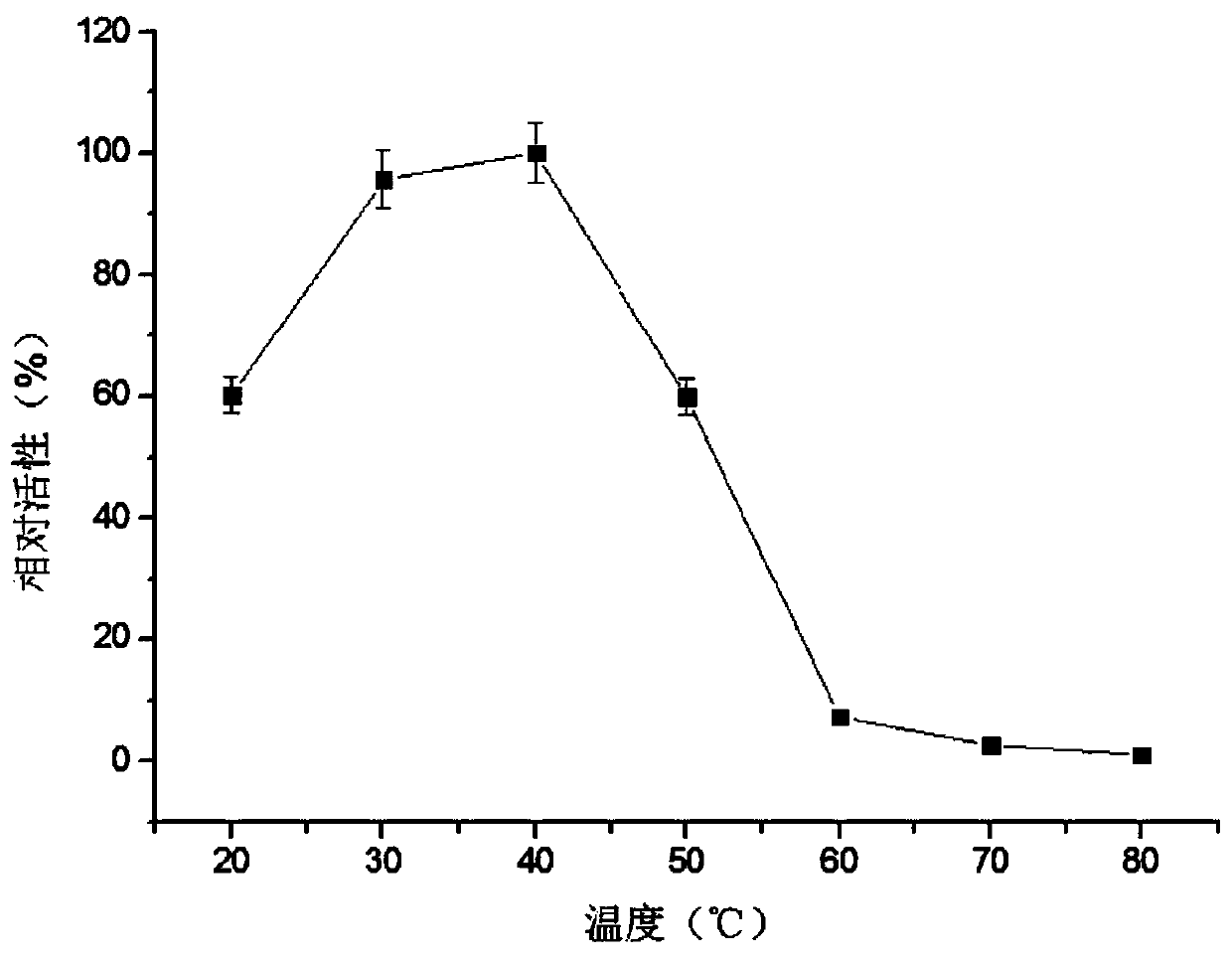
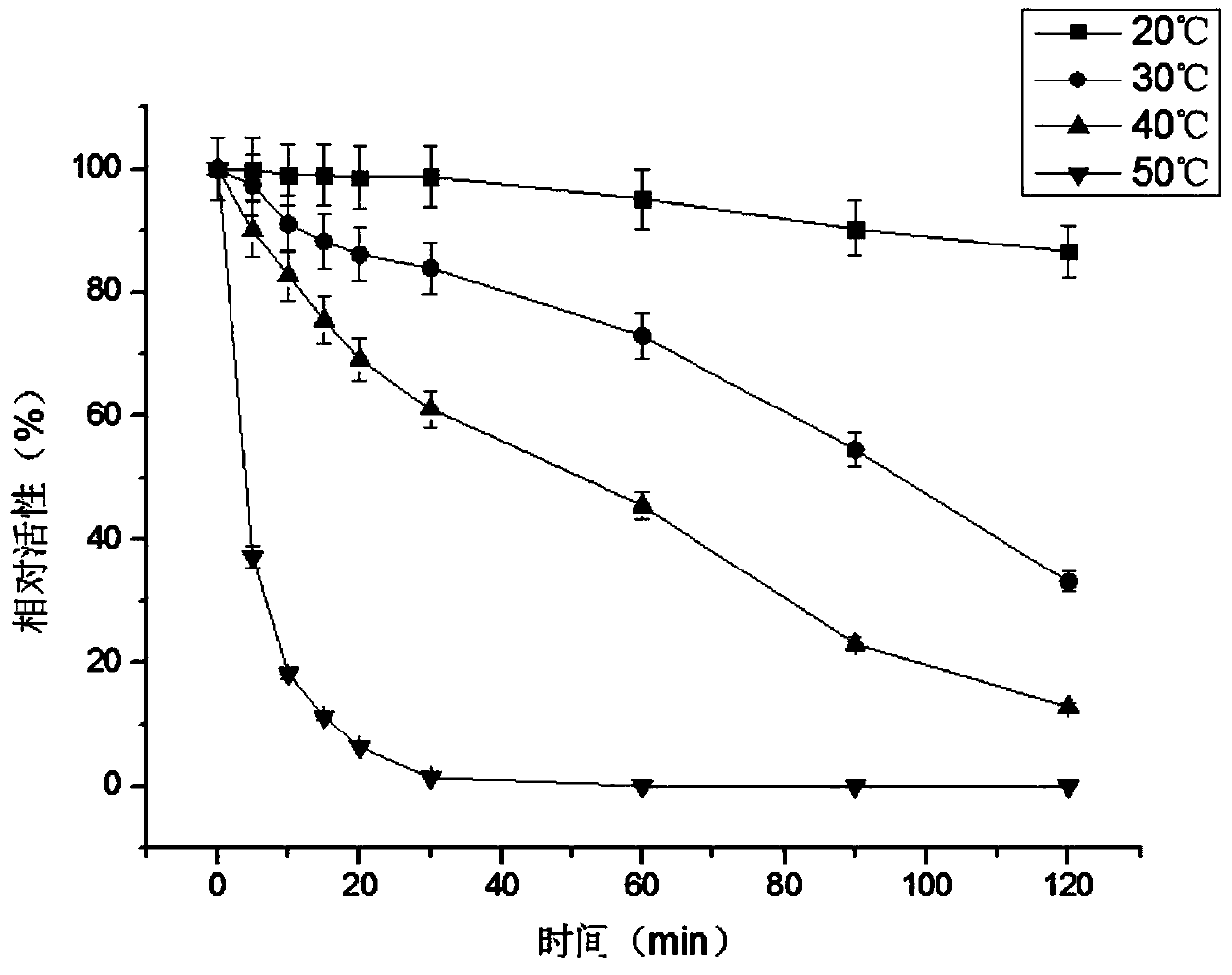
Method used
Image
Examples
Embodiment 1
[0039] Embodiment 1 obtains β-glucosidase gene
[0040] This example illustrates the method for obtaining the β-glucosidase gene of the present invention. Specific steps are as follows:
[0041] (1) Sample collection: Soil samples were collected near the small garden beside the east playground of Nanjing University of Technology.
[0042] (2) Extraction of total soil DNA: Weigh 1 g of soil sample, add 1 mL of 0.1mol / mL, pH 8.0 phosphate buffer and glass beads, shake for 1 min, add 5 mg of lysozyme to make the final concentration of lysozyme 2.5 mg / mL, Shake at room temperature for 15 minutes, place in refrigerator for 30 minutes, add 125 μL of 20% SDS for shaking treatment for 15 minutes, centrifuge, add phenol (1:1 volume) for extraction once, chloroform-isoamyl alcohol (1:1 volume) for extraction twice, add 0.6 volume of isopropanol, placed at room temperature for 1 hour, centrifuged, washed with 70% ethanol, and dissolved in 30 μL TE.
[0043] (3) Construction of a genom...
Embodiment 2
[0052] The amplification of embodiment 2β-glucosidase gene
[0053] This example illustrates the specific process of β-glucosidase gene amplification.
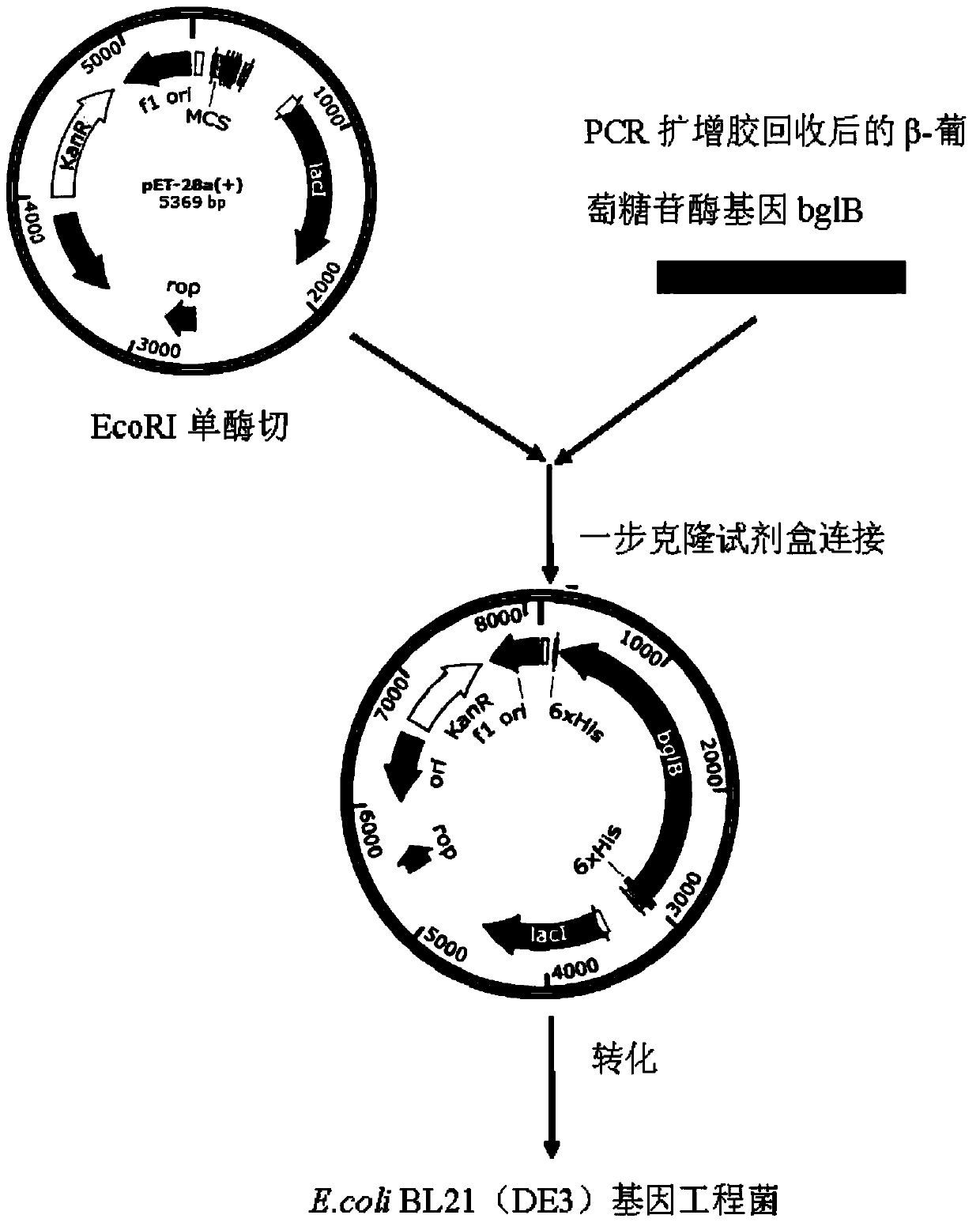
[0054] see figure 1 , using PCR to clone the β-glucosidase gene of the present invention. The designed primers are as follows:
[0055] Upstream primer P1: Tm=66.7, 45mer; the primer sequence is as follows:
[0056] CAAATGGGTCGCGGATCCGAATTCATGAAAGTAAAATCAACATGG;
[0057] Downstream primer P2: Tm=68.7, 52mer; primer sequence is as follows:
[0058] TTGTCGACGGAGCTCGAATTCTTACTTTTTTTGCCTTTTCTGTAGAGGTTGCC;
[0059] Use PCR to amplify the β-glucosidase gene. In a 50 μL reaction system, the amount of the two primers P1 and P2 added is 1 μL. The amplification conditions are: 94°C preheating for 10 minutes; 94°C for 45s; 55°C for 45s ; 72°C, 2.5min; 72°C, 10min, three steps in the middle, a total of 30 cycles, the DNA polymerase used is 2×Phanta @ Master Mix enzyme (Novazyme, China).
[0060] After the PCR was finished, the fra...
Embodiment 3
[0061] Embodiment 3β-glucosidase gene is expressed in escherichia coli
[0062] The above sequence of SEQ ID NO: 1 was directly connected to the prokaryotic expression vector pET-28a(+), and reacted at 37°C for 30 minutes. This vector was transformed into competent DH5α. A single colony was obtained by coating the bacterial solution on a solid LB medium (mass fraction formula: 2% peptone, 1% yeast powder, 1% NaCl, 1-2% agar) containing 30 μg / mL ampicillin. Pick a single colony and culture it overnight in a 5mL liquid LB test tube, extract the plasmid, and perform single and double enzyme digestion verification. Select and verify that the correct plasmid is transformed into Escherichia coli E.coli BL21(DE3). A single colony was obtained after the bacterial liquid was coated with solid LB medium containing 30 μg / mL ampicillin and cultivated. Then pick a single colony and culture it overnight in a 5mL liquid LB test tube, extract the plasmid, perform single and double enzyme d...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com