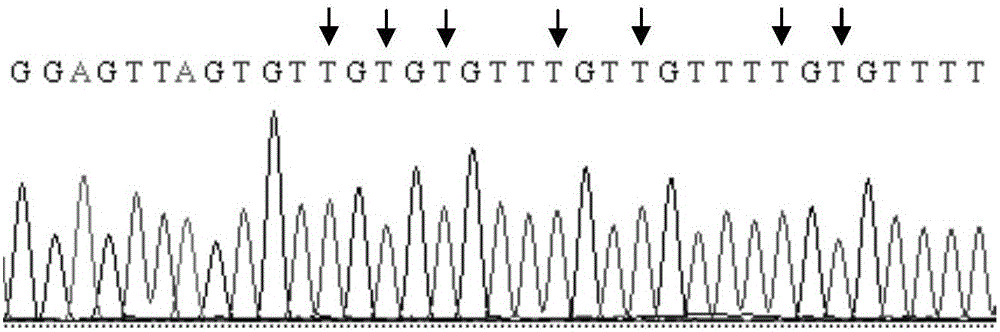
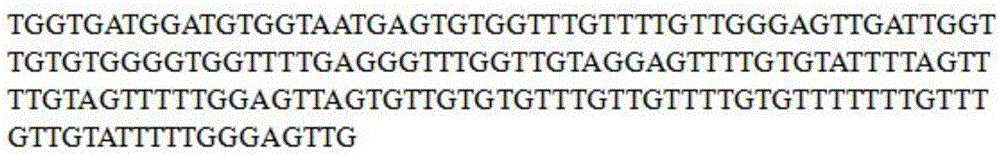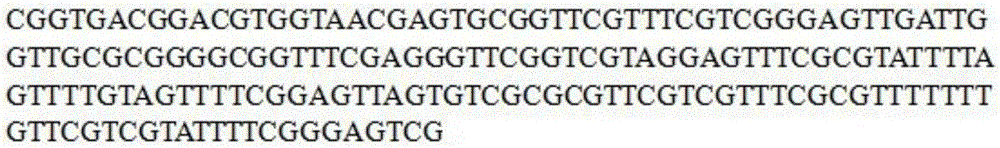SFRP1 gene promoter methylation detection primers and detection method thereof
A methylation and promoter technology, applied in the fields of life science and biology, can solve the problems of high instrument requirements, high probe cost, cumbersome operation, etc., and achieve cost saving, good detection stability, and high detection rate Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] The SFRP1 gene promoter region CG island methylation detection kit includes a pair of specific degenerate primers SEQ NO 1 and SEQ NO 2. At the same time, this primer is a sequencing primer, and its base sequence is as follows:
[0035] SEQ NO 1: 5'-CGGTGACGGACGTGGTAAYGAG-3';
[0036] SEQ NO 2: 5'-CGACTCCCGAAAATACGACRAACA-3';
[0037] SFRP1 gene promoter region CG island methylation detection kit also includes the following reagents:
[0038] (1) Sulfite treatment reagents: Bisulfite Mix, RNase free water, DNA Protect Buffer;
[0039] (2) PCR amplification reagents: 10*PCR Buffer, 2mM dNTP, 5*Q Solution Buffer, 5U / ul Taq enzyme, 10uM amplification primers (SEQ NO1, SEQ NO 2) and sterilized water;
[0040] (3) Enzyme purification reagents: EXOI, CIP;
[0041] (4) Sequencing reagents: Bigdye Terminator V3.1, absolute ethanol, EDTA, and sequencing primers (SEQ NO 1, SEQ NO 2).
[0042] (5) Positive quality control product: the methylated DNA of the CG island in the promo...
Embodiment 2
[0044] DNA extraction reagents and methods.
[0045] Take 5-8 paraffin sections (5-10μm thick, 1×1cm2 size), collect the paraffin-embedded tissue into a 1.5mL centrifuge tube, and centrifuge briefly; add 1mL tissue clear solution, shake fully, centrifuge at 13200rpm for 1min, remove the upper Supernatant solution: add 0.5mL tissue clear solution, shake fully, centrifuge at 13200rpm for 1min, remove supernatant; add 1mL ethanol (96-100%), shake vigorously, centrifuge at room temperature for 2min at 13200rpm, remove supernatant; open the lid, Incubate at room temperature (15-25°C) or in a metal bath at 37°C until the remaining ethanol is completely evaporated; take 180 μL Buffer ATL to resuspend the tissue in the tube, then add 20 μL proteinase K, and shake again; place the centrifuge tube containing the mixture in On a metal bath at 56°C, incubate for 1 hour, invert and mix 3 times until the sample is completely dissolved; transfer the centrifuge tube to a metal bath at 90°C, a...
Embodiment 3
[0047] The sulfite treatment reagents and methods are as follows.
[0048] Operation steps: dilute the genomic DNA extracted in Example 2 and 1.5ml EP tube to 50ul with double distilled water, add 5.5ul freshly prepared 3M NaOH solution, and bathe in water at 42°C for 30min; prepare the following solution during the water bath: 10mM hydroquinone solution (Hydryquinone) : Dissolve 55mg hydroquinone in 50ml water; 3.6mol / LNaHSO 3 : Take 18.8g NaHSO 3 Add 40ml of water, adjust the pH of the solution to 5.0 with 3M NaOH solution, and finally dilute to 50ml. Add 30ul 10mM hydroquinone solution and 520ul3.6mol / LNaHSO 3 Put the EP tube into the solution after the water bath, wrap the EP tube with aluminum foil to avoid light, gently invert the solution, add 100ul paraffin oil to the tube, and centrifuge briefly to cover the liquid surface with paraffin oil to prevent water evaporation during the water bath and limit oxidation by 50 ℃ Protect from light and water bath for 16 hours;...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com