System and method for in vitro detection of activity of Ub (ubiquitin)-conjugating enzyme
A ubiquitin-conjugating enzyme, in vitro detection technology, applied in microorganism-based methods, introduction of foreign genetic material using carriers, chemical instruments and methods, etc. Achieve the effect of reducing false negatives and reducing protein activity loss
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0071] Example 1. Establishment of ubiquitin-conjugating enzyme activity detection system
[0072] The main components and steps of establishing the ubiquitin-conjugating enzyme activity detection system are as follows:
[0073] 1. The Escherichia coli strain used for protein expression is BL21(DE3); prepare BL21(DE3) competent cells according to the method in the specific embodiment (B).
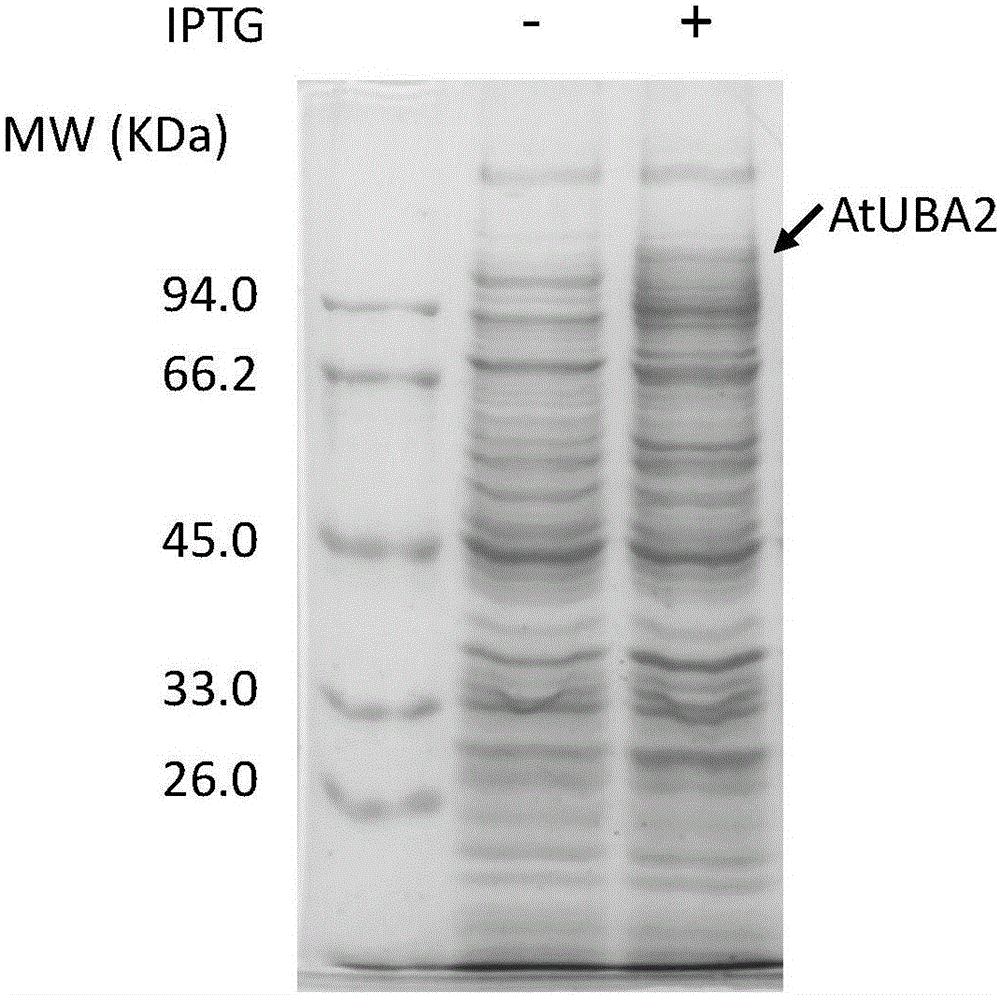
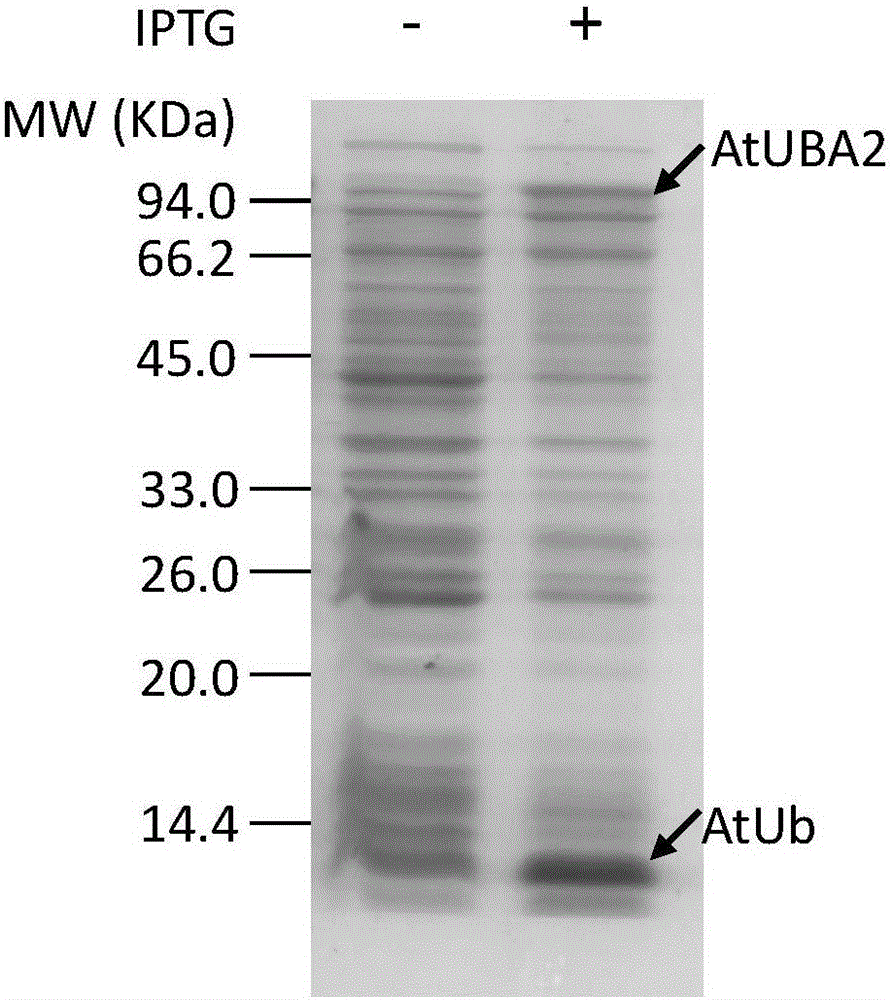
[0074] 2. Arabidopsis AtE1 is selected from Arabidopsis AtUBA2, and the protein expression vector comes from references: Qingzhen Zhao, Miaomiao Tian, Qiuling Li, Feng Cui, Lijing Liu; Bojiao Yin, Qi Xie. A plant-specific in vitroubiqutination analysis system. The Plant Journal, 2013, Vol. 74, pp. 524-533. The protein expression vector used was pGEX-6P-1 (Novagen).
[0075] 3. Construction of the Arabidopsis thaliana AtUb protein expression vector: the protein expression vector is pACYCDuet (Novagen); the sequence of the wild-type monomeric ubiquitin protein is derived from PET28a-AtUb ...
Embodiment 2
[0082] Example 2. Detection of Arabidopsis AtUBC10 ubiquitin-conjugating enzyme activity
[0083] 1. Arabidopsis AtUBC10 is an E2 protein known to have ubiquitin-conjugating enzyme activity (references: Qingzhen Zhao, Miaomiao Tian, Qiuling Li, Feng Cui, Lijing Liu; Bojiao Yin, Qi Xie. A plant-specific in vitroubiqutination analysis system.The Plant Journal, 2013, volume 74, pages 524-533), we use this protein to verify whether the ubiquitin-conjugating enzyme activity detection system in this patent works.
[0084] 2. Construction of Arabidopsis thaliana ubiquitin-conjugating enzyme AtUBC10 protein expression vector: We used the pCDFDuet (Novagen) plasmid, which is different from the replication origin of pGEX-6P-1 and pACYCDuet, as the prokaryotic expression vector of E2 protein; we used EcoRI and SalI to construct AtUBC10 On the pCDFDuet carrier, and add a Flag tag at the carboxyl end of AtUBC10 to facilitate Western blot detection (primers used are AtUBC10-F and AtUBC10-...
Embodiment 3
[0087] Example 3. Peanut AhUBC34 Ubiquitin Conjugating Enzyme Activity Detection
[0088] 1. Construction of the peanut AhUBC34 prokaryotic protein expression vector: The peanut AhUBC34 is derived from the transcriptome sequencing results of the cultivar Luhua 14, and its amino acid sequence is shown in SEQ NO.1 in the sequence table; Solubility of the protein, we removed the transmembrane domain of AhUBC34, constructed AhUBC34 on the pCDFDuet vector with EcoRI and SalI, and passed primers (the primers used are AhUBC34-F and AhUBC34ΔTM-R as shown in the sequence table SEQ NO.5-6 ) amplified and added a Flag tag to the C-terminus of the AhUBC34 protein to facilitate Western blot detection, thereby constructing the vector plasmid pCDFDuet-AhUBC34 capable of expressing the E2 protein in vitro. Transfer pCDFDuet-AhUBC34 into BL21(DE3) cells according to the method described in the specific embodiment (C), select a single clone for culture, and induce expression according to the me...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com