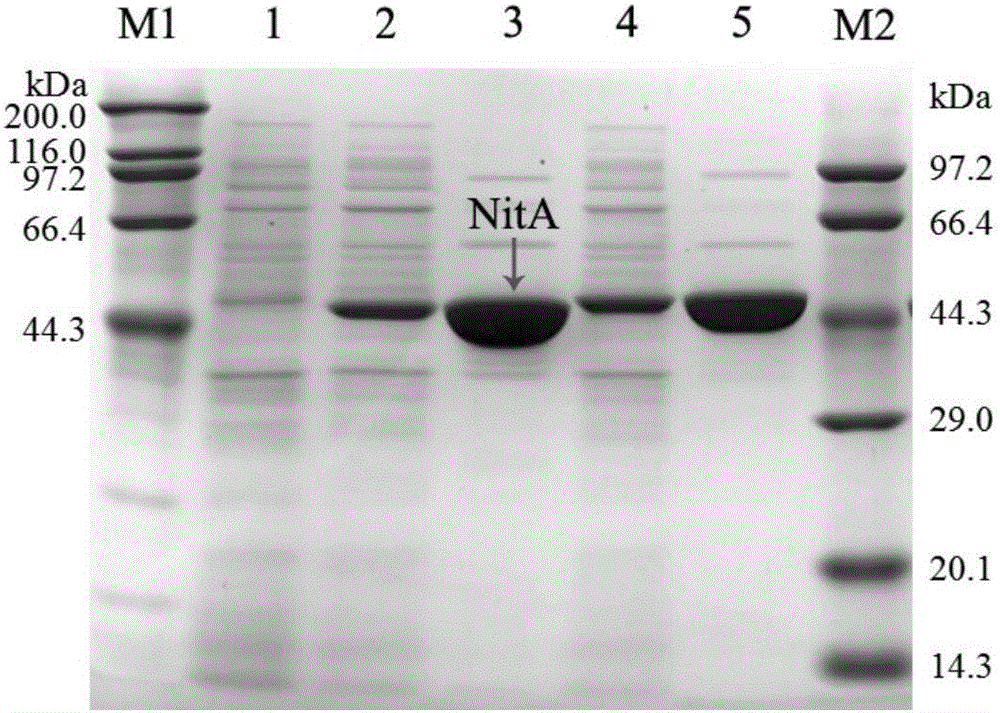
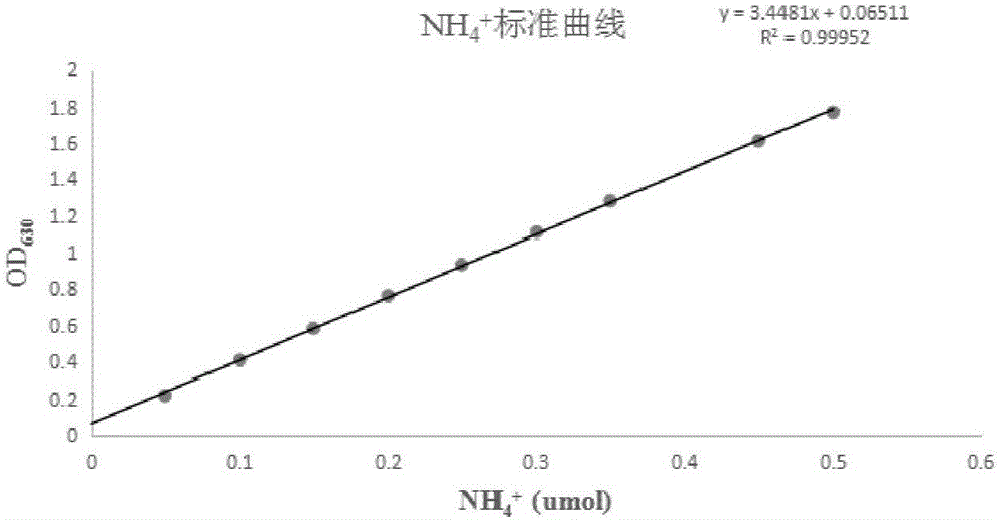
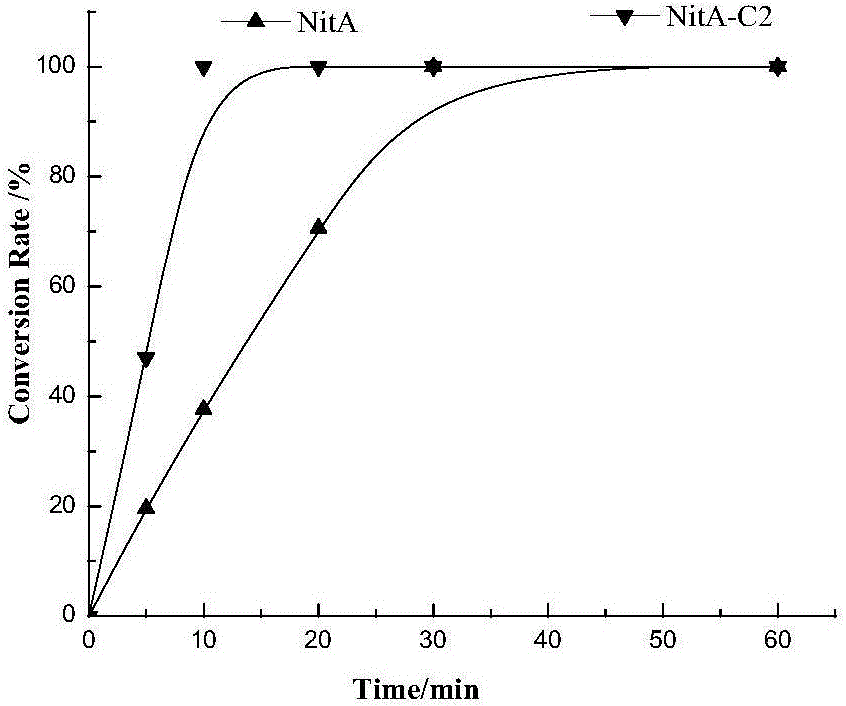
Nitrilase mutant and application thereof in preparation of nicotinic acid
A nitrilase and mutant technology, applied in the fields of molecular biology and biology, can solve the problems of complex reaction process, serious environmental pollution and high energy consumption, and achieve the effect of high enzymatic reaction rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0017] Example 1. Construction of Prototype Nitrilase Recombinant Bacteria E.coli BL21pET28a-nitA
[0018] Using the synthetic gene SEQ ID NO.1 (NCBI GenBank No.DQ444267, synthesized by GenScript) as a template, according to the gene sequence of SEQ ID NO.1 and the polyclonal enzyme of Escherichia coli expression vector pET-28a (+) Cut site design primers.
[0019] Upstream primer: P1: 5'-GGTGCCGCGCGGCAGCCATATGGTTTCGTATAACAGCA-3'.
[0020] Downstream primer: P2: 5'-GCGGCCGCAAGCTTGTCGACCTTTGCTGGGACCGGTTCT-3'.
[0021] The PCR reaction conditions are:
[0022]
[0023] The reaction conditions of PCR were: cover temperature at 99°C; pre-denaturation at 94°C for 10 min; denaturation at 94°C for 45 s, annealing at 50°C for 45 s, and extension at 72°C for 2 min, a total of 32 cycles. The PCR product was verified using 0.8% agarose gel, and after the verification was correct, the fragment was cut using the Biomiga DNA gel recovery kit to recover the fragment.
[0024] Escheric...
Embodiment 2
[0028] Embodiment two, the construction of mutant bacteria
[0029] Using the synthetic gene SEQ ID NO.1 in Example 1 as a template, primers were designed according to the gene sequence of SEQ ID NO.1 and the nucleic acid sequence of the mutation site.
[0030]
[0031] The recombinant plasmid pET-28a(+)-Nit containing the target gene fragment was used as a template, and the template was amplified according to the method of overlap extension PCR, and the principle of the method was the same as in Example 1. The whole amplification process is carried out in two steps. First, primers P1 and F168V-F were added respectively, and the template was amplified with P2 and F168V-R to obtain two DNA sequences. Then add the two sequences into a PCR system, add primers P1 and P2, and continue PCR amplification to obtain the full-length F168V mutant DNA sequence. On the basis of the mutant at position 168, the amino acid at position 192 was mutated according to the above method to obta...
Embodiment 3
[0033] Embodiment three, the expression of nitrilase gene
[0034] Pick a single colony and insert it into a 5ml liquid LB medium test tube, and the medium contains 0.1g / L kanamycin. Culture overnight at 37°C, 200 rpm, with shaking. The next day, the liquid seeds in the test tube were transferred to fresh LB liquid medium containing 0.1g / L kanamycin with an inoculum size of 3% by volume, and cultivated at 37°C until the OD of the bacteria 600 When it is about 0.4-0.8, add the inducer IPTG (final concentration is 0.1mM) to the above LB liquid medium, induce for 20h at 25°C, then centrifuge the fermentation broth at 4°C and 8000rpm for 10min, and collect the wet cells. The bacterial cells were washed twice with PBS buffer solution of pH=7.0, and the bacterial cells were collected.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com