Gene, expression vector, bacterial strain and application of a kind of sterol lipase for papermaking
An expression vector, a technology of sterol lipase, applied in the field of bioengineering, can solve the problems of paper breakage, reduced paper machine speed, complex composition, etc., and achieve the effect of stable operation efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036]Embodiment 1 synthetic sterol esterase gene (CHE)
[0037] The sterol esterase (CHE) gene sequence and protein structure information of S.lavendulae H646-SY2 was obtained from the National Center for Biotechnology Information (NCBI, http: / / www.ncbi.nlm.nih.gov / ) and the Protein Database (PDB, http: / / www.rcsb.org / pdb / search / advSearch.do).
[0038] According to the CHE gene sequence published on NCBI, the sequence was optimized, and all the codons preferred by Pichia pastoris were replaced, and then the optimized gene sequence was used with DNAWORKS software ( http: / / mcl1.ncifcrf.gov / lukowski.html ), design primers for whole gene synthesis, and design 2 primers, wherein 2 primers introduce EcoRI and NotI enzyme cutting sites and enzyme cutting protection bases respectively, and the primer sequences are:
[0039] CHE-F: 5′-CCG GAATTC ATGTCCTCCCAAGTT-3′; (the horizontal line is the EcoRI restriction site)
[0040] CHE-R: 5′-ATAAGAAT GCGGCCGC TTAATGG-3'; (the horiz...
Embodiment 2
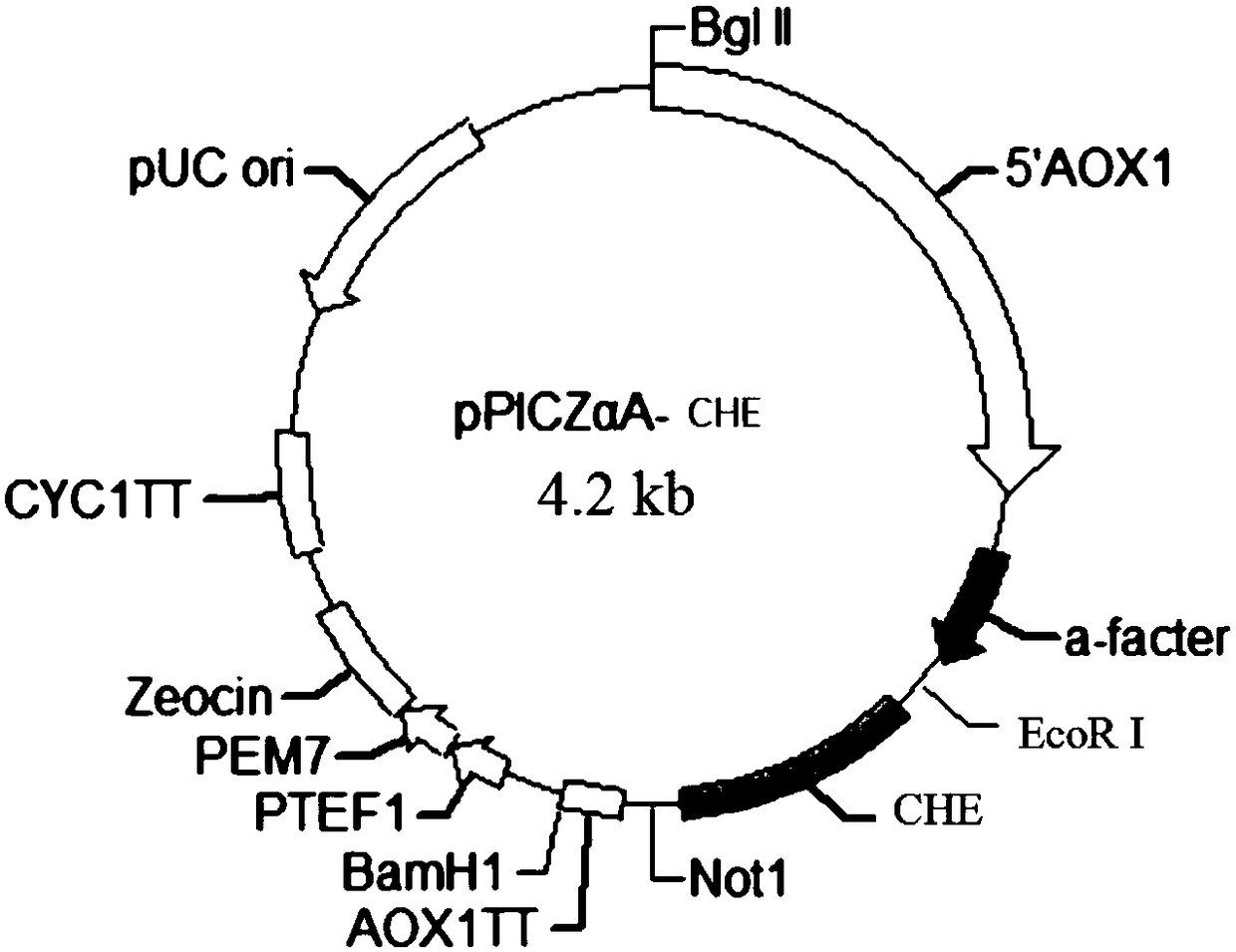
[0042] Embodiment 2 prepares the plasmid containing CHE gene
[0043] The PCR product obtained in Example 1 was electrophoresed, the gel was cut to recover the target band at about 700bp, the recovered product was digested with EcoRI and NotI, and the expression vector pPICZαA was also digested with EcoRI and NotI and recovered from the gel with T4DNA Ligase ligation. The 10 μL volume reaction system is as follows: pPICZαA 1 μL (50ng), add 6 μL of fully synthetic gene product, 1 μL of 10× Buffer containing ATP, T4 DNA ligase 1 μL, add ddH 2 O to make up to 10 μL. Slightly centrifuged, connected in a water bath at 16°C overnight, and transformed into E.coli Top10 (invitrogen), and positive transformants were screened on LB plates containing Zeocin (bleomycin, 25 μg / mL). Randomly pick a certain amount of transformants, prepare a small amount of plasmids, and perform electrophoresis analysis after double digestion with restriction endonucleases EcoRI and NotI. It is estimated ...
Embodiment 3
[0044] Example 3 High expression of sterol lipase CHE in Pichia pastoris
[0045] The pPICZαA-CHE of Example 2 that had been completely linearized by Sac I was transformed into Pichiapastoris X33 (invitrogen) by the LiCl method. The transformants were spread on MD plates and cultured at 30°C for 2 days. Transformants on the MD plate were inoculated on YPD plates containing Zeocin 100 μg / mL, 200 μg / mL, 300 μg / mL, 400 μg / mL, and 500 μg / mL, and cultured at 30°C for 3 days. Single clones were picked from the transformants appearing on the Zeocin-YPD plate with a higher concentration. Extract yeast genomic DNA as a template according to the Invitrogen operating guide, and use the PCR primers of the target gene sequence for yeast genome PCR identification. The total reaction volume is 20 μL, and the amount of Taq enzyme is 2U. Take 2 μL of PCR products for 0.8% agarose gel electrophoresis identification.
[0046] The PCR amplification product was detected by 1% agarose gel electro...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com