Method for identifying balanced translocation breaking point and balanced translocation carrying state of embryo
A technique for balancing translocations and breakpoints, which is applied in genome sequence analysis and the biological field, and can solve problems such as inability to be used as a general detection technique, unstable probe hybridization efficiency, and time-consuming
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
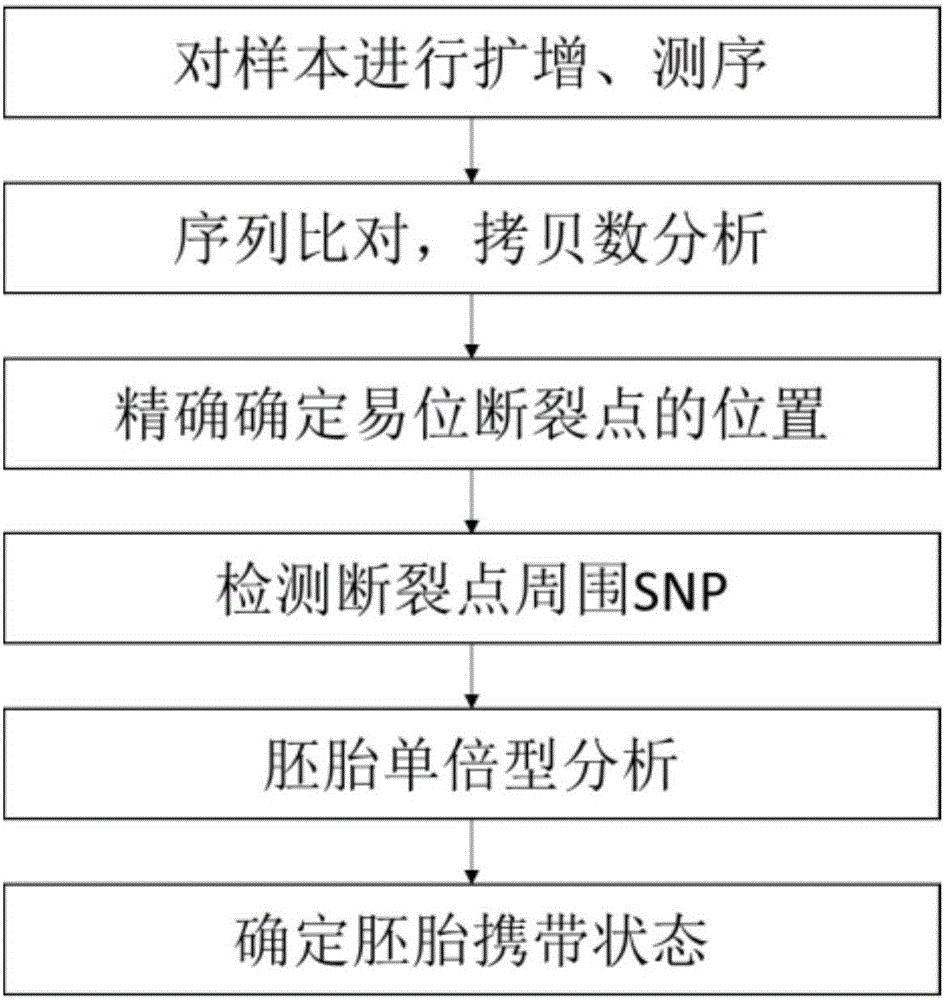
[0084] Example 1 Determination of Balanced Translocation Breakpoint
[0085] (1) Obtain embryo samples to be tested and parental DNA
[0086] After IVF, this family obtained 8 embryos, the embryos developed to the blastocyst stage, and 3-5 trophectoderm cells were removed from each embryo biopsy.
[0087] (2) Amplify and sequence the samples
[0088] The single cell amplification adopts the multi-annealing circular cycle amplification technology single cell whole genome amplification kit YK001A of Shanghai Yikang Medical Laboratory Co., Ltd., and operates according to the instructions provided by Shanghai Yikang Medical Laboratory Co., Ltd., and the embryo biopsy Cells undergo whole genome amplification.
[0089] Sequencing was performed using the HiSeq2500 high-throughput sequencing platform of Illumina, and operated according to the instructions provided by Illumina. The sequencing type was single-end (Single End) sequencing, the sequencing length was 50 bp, and the seque...
Embodiment 2
[0105] Example 2 Determination of Carrying State of Balanced Translocation of Embryo
[0106] (1') the determined equilibrium embryo equilibrium translocation breakpoint obtained from Example 1;
[0107] (2') Detection of SNPs around the breakpoint
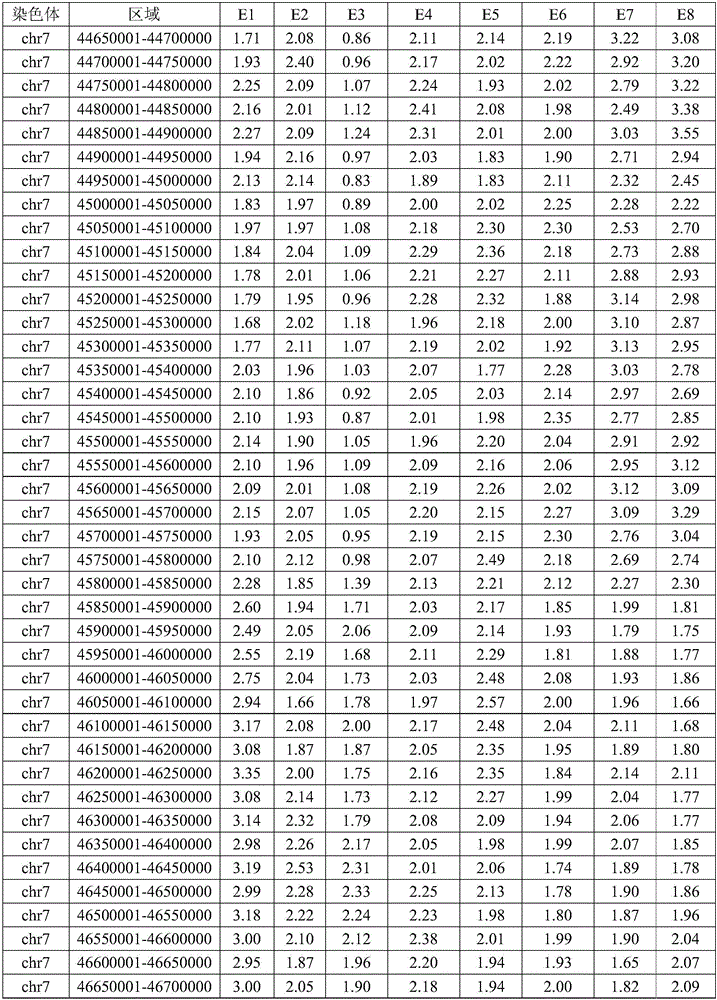
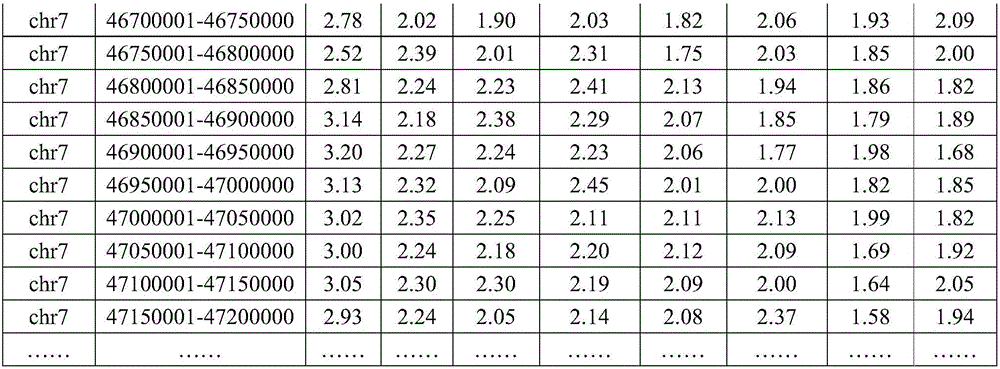
[0108] at a distance of 1 x 10 from chr7 breakpoint 6 In the range of bp, 61 SNP sites of embryonic parents and embryos were detected respectively; at a distance of 1×10 from chr16 breakpoint 6 In the range of bp, 63 SNP sites of embryo parents and embryos were detected respectively. The detection method of the SNP site is to design primers to perform next-generation sequencing on the amplicon. The method of determining the genotype is next-generation sequencing, and the analysis software is used to determine the SNP genotype for SAMtools. The SNP results upstream and downstream of chr7 and chr16 breakpoints in this family are shown in Table 3 and Table 4.
[0109]Table 3 SNP genotypes upstream and downstream of chr7 breakpoi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com