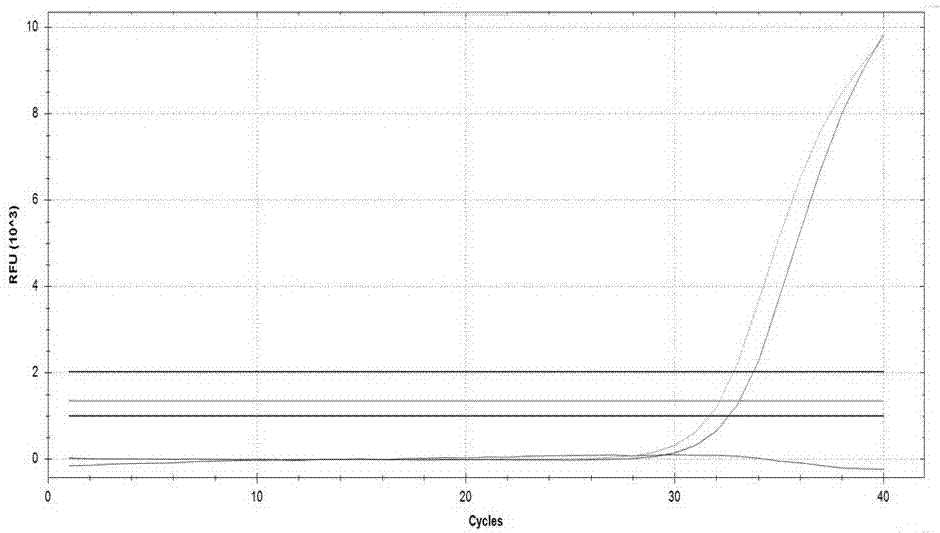
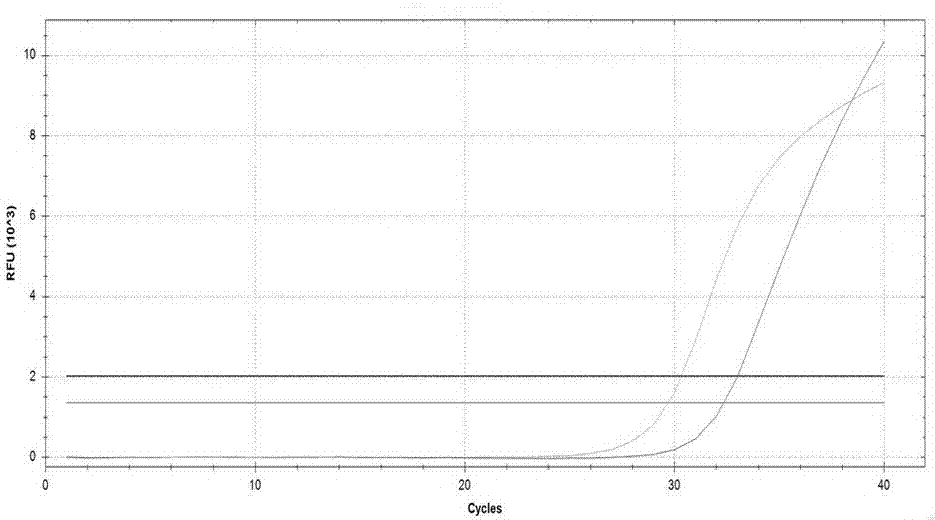
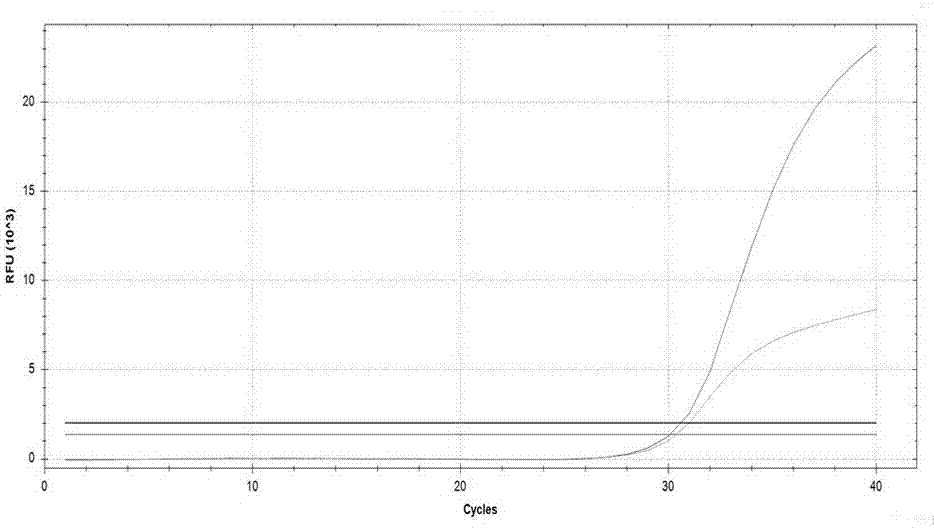
Kit used for detecting helicobacter pylori drug resistance gene mutation via multiple fluorescent PCR
A Helicobacter pylori, multiple fluorescence technology, applied in the field of fluorescence quantitative PCR, can solve the problems of complicated operation of sequencing method, long detection period, complicated steps, etc., and achieve the effects of improving the success rate of treatment, reducing the cost of testing, and reducing the economic burden.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] 1. Reagent preparation
[0044] (1) Preparation of clarithromycin resistance reaction buffer
[0045] Take a 10mL volumetric flask and add 0.5mol / L Trizma ® HCl 80μL, 0.5mol / L Trizma ® Base 720μL, 100mmol / L MgCl 2 200 μL, 5mol / L KCl 160 μL, formamide 300 μL, 1mol / L ammonium sulfate 400 μL, 100 mmol / L dNTPs 90 μL, 100 μmol / L CLW upstream primer 33.3 μL, 100 μmol / L 2142, 2143, 2182 downstream primers each 33.3 µL, 10 µL of 100 µmol / L CLW probe, 20 µL of 100 µmol / L internal control upstream primer, 20 µL of 100 µmol / L internal control downstream primer, 5 µL of 100 µmol / L internal control probe. Make up the volume to 10mL with sterilized ultrapure water, mix well, dispense 1.5mL / tube into centrifuge tubes, and store at -20°C for later use.
[0046] (2) Preparation of Tetracycline Resistance Reaction Buffer
[0047] Take a 10mL volumetric flask and add 0.5mol / L Trizma ® HCl 80μL, 0.5mol / L Trizma ® Base 720μL, 100mmol / L MgCl 2 200 μL, 5mol / L KCl 160 μL, formamide...
Embodiment 2
[0075] 1. Reagent specificity verification
[0076] (1) Experimental samples
[0077] Six specific samples were taken to verify the specificity of the reagents, namely Escherichia coli, Campylobacter jejuni, Salmonella, Enterovirus 71, Coxsackievirus 16, and Rotavirus.
[0078] (2) Experimental process
[0079]The reagents described in this method were used to detect the above six specific samples, and the test results were analyzed to verify the specificity of the reagents.
[0080] (3) Experimental results
[0081] The test results of 6 specific samples were all negative, indicating that the reagent has good specificity and no cross-reaction. The specific results are shown in the table below:
[0082] Specific test results
[0083]
[0084] 2. Reagent precision verification
[0085] (1) Experimental samples
[0086] The precision of the reagents was verified by the HP drug-resistant positive control.
[0087] (2) Experimental process
[0088] Repeat the detection...
Embodiment 3
[0101] Embodiment 3: Detect the result of 60 routine clinical samples
[0102] 1. According to the preparation method shown in Example 1, the relevant components of the kit were prepared and stored at -20°C for later use.
[0103] 2. Obtained 60 cases of clinical gastric mucosal paraffin section samples from the Fuzhou General Hospital of Nanjing Military Region, and used the paraffin tissue nucleic acid extraction reagent (magnetic bead method) of Jiaxing Yakangbo Beinan Biotechnology Co., Ltd. to extract the genomic DNA of 60 cases of clinical samples. The purity of the DNA samples was tested by a photometer, and the OD260 / OD280 of 60 samples were all between 1.6 and 2.0.
[0104] 3. According to the steps shown in Example 1, add DNA samples and perform detection on a fluorescent quantitative PCR instrument. The instrument used this time is Bio-rad CFX96.
[0105] 4. According to the judgment standard shown in Example 1, the results are interpreted and counted, and the resu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com