Method for synthesizing L-2-piperidinecarboxylic acid through whole-cell catalysis
A pipecolinic acid and whole-cell technology, which is applied in the field of whole-cell catalytic synthesis of L-2-piperidinecarboxylic acid, can solve the problems of low pipA enzyme activity, low substrate concentration, and large amount of whole cells, and achieve enzyme activity High efficiency, short reaction time, and high ee value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
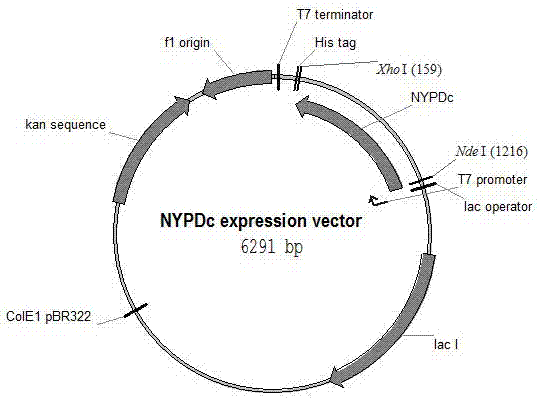
[0037] Arenimonas donghaensis DSM 18148 was placed in LB medium, cultured at 30°C and 180 rpm for 3-5 days, the precipitate was collected by centrifugation, and the DNA of Arenimonas donghaensis DSM 18148 was extracted and purified with the DNA extraction and purification kit QiAamp Kit (Qiagen, Germany).
[0038] The Genomic DNA of Arenimonas donghaensis DSM 18148 was amplified by PCR with Pfu high-fidelity enzyme, and the primers used were
[0039] NYPD-F 5' ATGACCATGACCCAGCTCACCA 3'
[0040] NYPD-R 5' TCAGGCCGCCTGCTTGC 3'
[0041] Since the GC content of Arenimonas donghaensis DSM 18148 DNA fragment is close to 70%, betaine with a final concentration of 0.5M was added during amplification. Afterwards, the amplified fragment was treated with Taq polymerase at 72°C for 10 minutes, and base A was added to the 3' end of the DNA. Afterwards, it was connected to the pMD19T-simple (Takara Bao Biological Company, Beijing) cloning vector, and a single clone was picked and sent to ...
Embodiment 2
[0043] Due to the high GC content of the original DNA sequence, it is difficult to achieve high-efficiency transcription when cultured in host bacteria. Therefore, through the method of whole gene synthesis, the secondary structure and codon bias of the gene are adjusted to achieve high expression in Escherichia coli. Use Primer Premier (http: / / primer3.ut.ee / ) and OPTIMIZER (http: / / genomes.urv.es / OPTIMIZER / ) to design, and ensure that the Tm difference is controlled within 3°C, and the primer length is controlled within 50base , resulting in the following primers:
[0044]
[0045]
[0046] The above primers were synthesized, and the obtained primers were dissolved in double-distilled water, and then added to the following reaction system, so that the final concentration of each primer was 30 nM, and the final concentration of the first and last primers was 0.6 μM.
[0047] 2mM dNTP mix (2mM each dNTP)
5μl
10×Pfu buffer
5μl
Pfu DNA polymer...
Embodiment 3
[0051]Through the method of whole gene synthesis, the secondary structure and codon bias of the gene are adjusted to achieve high expression in Escherichia coli. Use Primer Premier (http: / / primer3.ut.ee / ) and OPTIMIZER (http: / / genomes.urv.es / OPTIMIZER / ) to design, and ensure that the Tm difference is controlled within 3°C, and the primer length is controlled within 50base , resulting in the following primers:
[0052]
[0053]
[0054] The above primers were synthesized, and the obtained primers were dissolved in double-distilled water, and then added to the following reaction system, so that the final concentration of each primer was 30 nM, and the final concentration of the first and last primers was 0.6 μM.
[0055] 2mM dNTP mix (2mM each dNTP)
5μl
10×Pfu buffer
5μl
Pfu DNA polymerase (10U / μl)
0.5μl
ddH2O
Bring the total volume of the reaction system to 50 μl
[0056] The prepared PCR reaction system was placed in ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com