A kind of improved transaminase, its encoding gene and genetic engineering bacteria expressing the enzyme
A technology of genetically engineered bacteria and transaminase, applied in genetic engineering, transferase, plant gene improvement, etc., can solve problems such as poor stereoselectivity, low yield, and expensive catalyst, and achieve good industrial application prospects and catalytic activity High, product chiral purity and high yield effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] The construction of embodiment 1 transaminase mutant coding gene
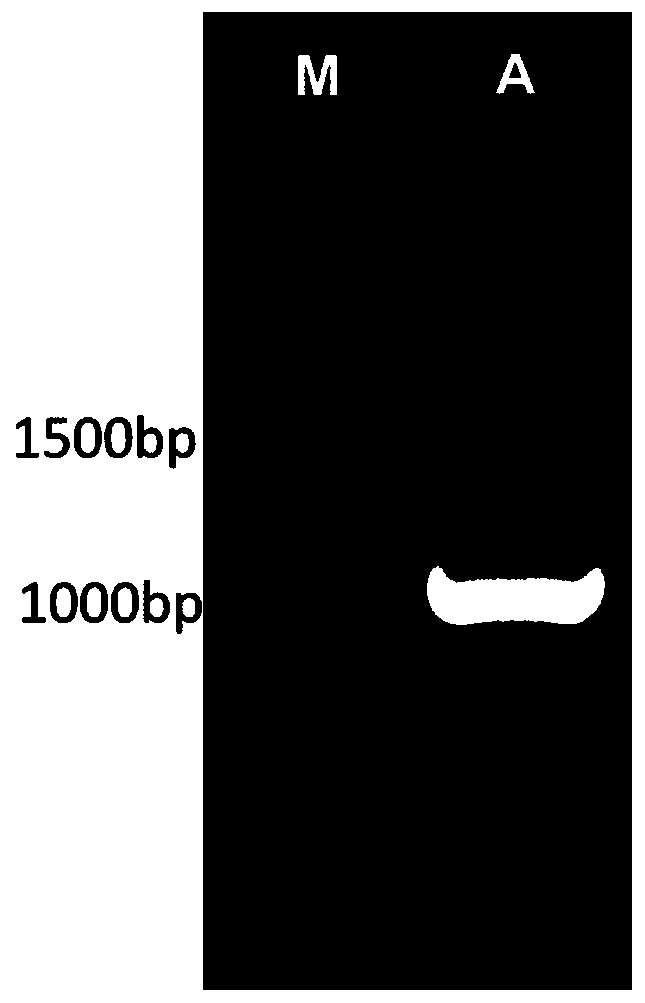
[0026] The synthetic sequence of the whole gene is shown in SEQ ID No.1, and two restriction sites, Nde I and Hind III, were selected and inserted into the pET-28a expression vector, and the obtained recombinant expression vector was named pET28a-AfAT. To construct the mutant library, we designed the following 8 primers, see Table 1 for details:
[0027] Table 1 PCR primer list
[0028]
[0029] Use pET28a-AfAT as a template and use the above primers for PCR amplification. The PCR system is: 10L of 5×PCR buffer, 4L of 2.5mmol / L dNTPs, 0.5L of DNA polymerase, 0.5L of pET28a-AfAT template (containing 0.1g of DNA template), ddH 2O is 31L, the AfAT-up upstream primer (SEQ ID No.5) and H53-down downstream primer (SEQ ID No.8), H53-up upstream primer (SEQ ID No.7) and E115- down downstream primer (SEQ ID No.10), E115-up upstream primer (SEQ ID No.9) and S214-down downstream primer (SEQ ID No.12), S214-u...
Embodiment 2
[0030] The construction of embodiment 2 transaminase mutant library
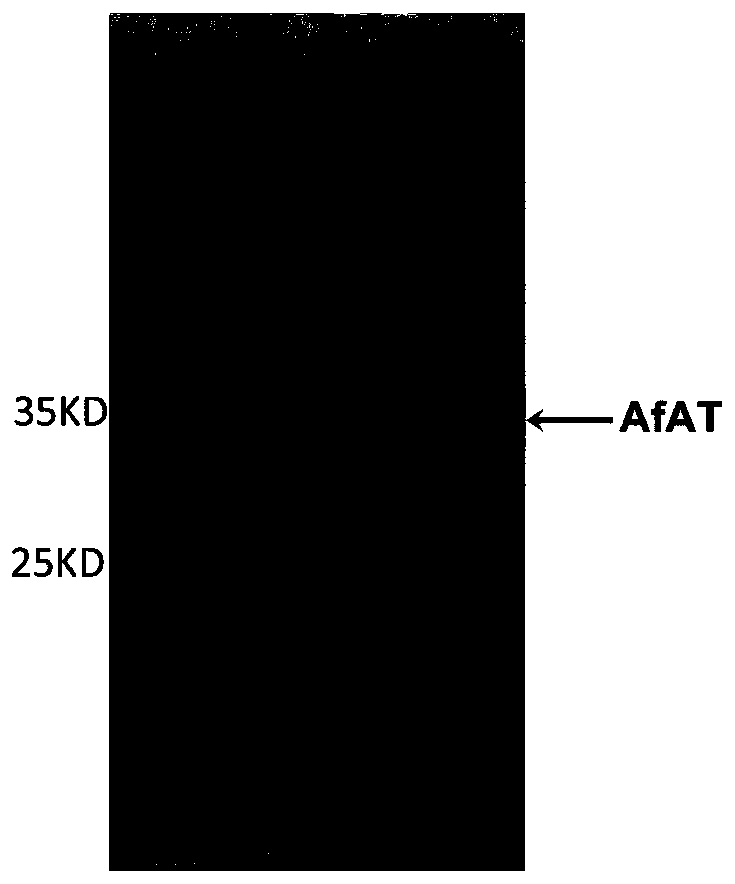
[0031] The above gene fragments and the vector pET-28a plasmid were subjected to enzyme digestion reactions (Nde I and Hind III, 37°C for 1 h), and the enzyme digested products were gel-cut and recovered, followed by ligation reaction (reaction overnight at 16°C), and transformed into Escherichia coli BL21(DE3) competent cells were screened with kanamycin to obtain positive single clones. Add 0.6 mL of LB medium (containing 50 g / L kanamycin) to each well of a 96-well plate, pick 93 positive clones and 3 BL21(DE3) / pET28a-AfAT clones from each 96-well plate as controls, and pick Twelve 96-well plates were taken and cultured in a shaker at 37°C for 16 hours, and this was the mutant library.
Embodiment 3
[0032] Expression, screening and identification of embodiment 3 transaminase mutants
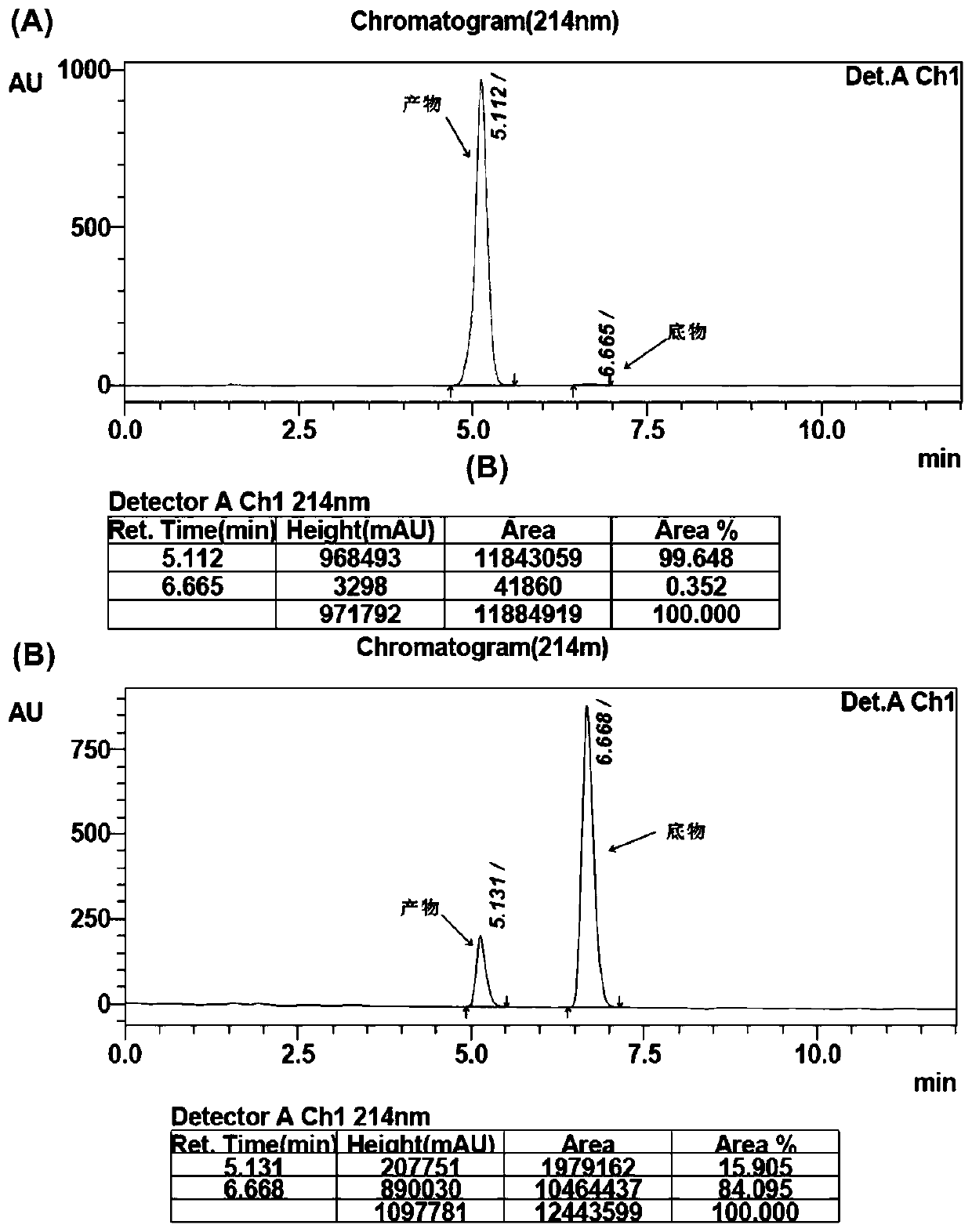
[0033] Transplant 200 L of the overnight mutant bacterial solution to a new 96-well plate, each well of which contains 1 mL of fresh LB medium, wherein the concentration of kanamycin is 50 g / L, and the concentration of IPTG is 1.0 mmol / L. Induce the culture at 25°C for about 20 hours, centrifuge to discard the supernatant, and collect the bacteria. Add 600L reaction solution to each well, including: 10g / L substrate p-methylacetophenone, 60g / L isopropylamine, 1mmol / L coenzyme pyridoxal phosphate (PLP), 100mmol / L potassium phosphate buffer (pH=8.0), after 24 hours of shaking reaction on a shaking table at 35° C., the reaction solution was analyzed by HPLC to detect the conversion rate of the mutant. Conversion rate detection conditions: Shimadzu2010AHT high performance liquid chromatography; Athena C18-WP (150 × 4.6mm, 5m) chromatographic column; wavelength 214nm; mobile phase A (H 2 O+0.1%T...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com