Preparation method of abyss sea cucumber sourced superoxide dismutase Cu, ZnSOD and application thereof
A kind of superoxide, dismutase technology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1 Obtaining of total RNA and cDNA of abyssal sea cucumber Paelopatides sp.:
[0032] The abyssal sea cucumber Paelopatides sp. was collected from the 6500m abyss of the Mariana Trench (latitude: 10°57.1693’N, longitude: 141°56.1719’E). After adding RNAlater to the sample for grinding, the sample was immediately stored in liquid nitrogen. After returning to the laboratory, take 50-100mg sample tissue, use RNase FREE experimental paper to blot the residual RNAlater, and put the tissue into a 2ml centrifuge tube. Add 1000ul QIAzol, and use Ruptor to stir for 20-40 seconds to fully break up the sample. Place the homogenate at room temperature for more than 10 min. The processed homogenate was subjected to total RNA extraction according to the relevant operations of the QIAGEN Total RNA Extraction Kit. The cDNA was synthesized by reverse transcription after detecting that the RNA was not degraded by electrophoresis. The reverse transcription from total RNA to cDNA...
Embodiment 2
[0034] Example 2 Obtaining and sequence analysis of Paelopatides sp. superoxide dismutase PaSOD gene:
[0035] Using the cDNA sequence obtained in Example 1 as a template, the PaSOD gene is amplified with primers F and R, wherein,
[0036] Primer F: CG GGATCCATGTCTGTCCACGCCGTTTGTGTATT, the nucleotide sequence of which is shown in SEQ ID NO.3;
[0037] Primer R:AA CTGCAG TATCTTTTTGATCCCAATTACA, the nucleotide sequence of which is shown in SEQ ID NO.4;
[0038] The underlined part GGATCC represents the restriction site of Bam H I, and CTGCAG represents the restriction site of Pst I.
[0039] The PCR reaction system is 50 μL, including 10 μL PrimeSTAR GXL Buffer, 4 μL dNTP, 1 μL each of F and R primers, 1 μL template cDNA, 2 μL GXL DNA Polymerase (TaKARa company), with H 2 O to bring the total volume to 50 μL. The PCR reaction conditions were: 98°C for 10s, 55°C for 15s, 68°C for 10s, 30 cycles, and stored at 4°C. PCR products were purified by tapping and pMD TM 18-T Ve...
Embodiment 3
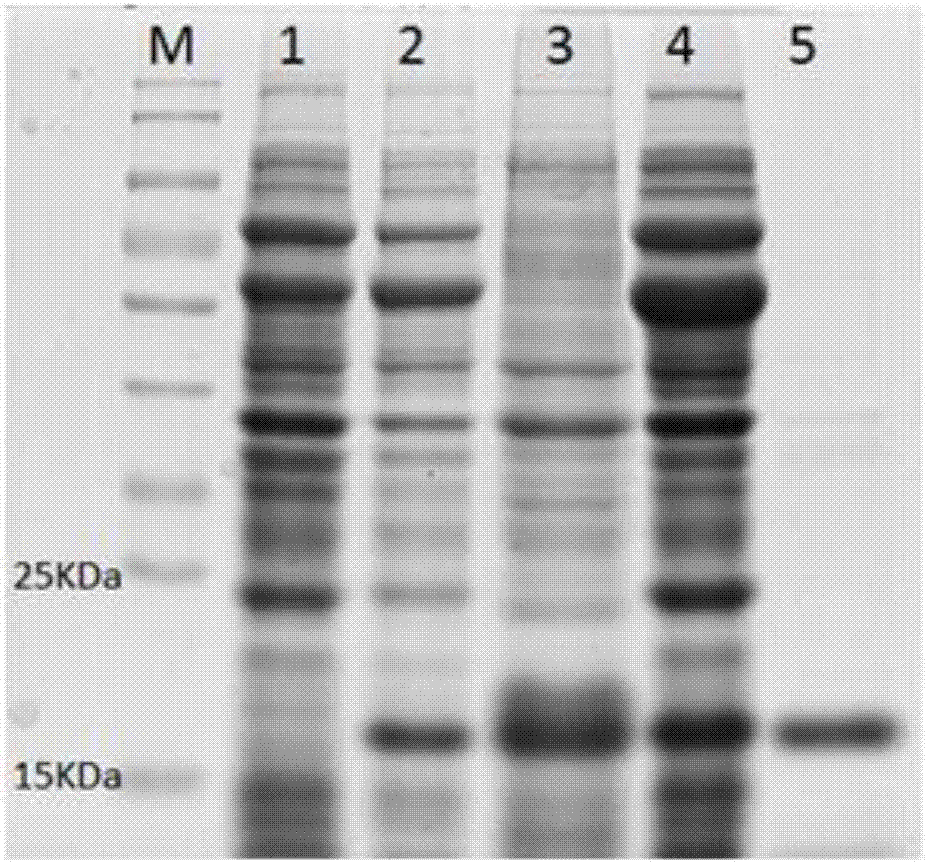
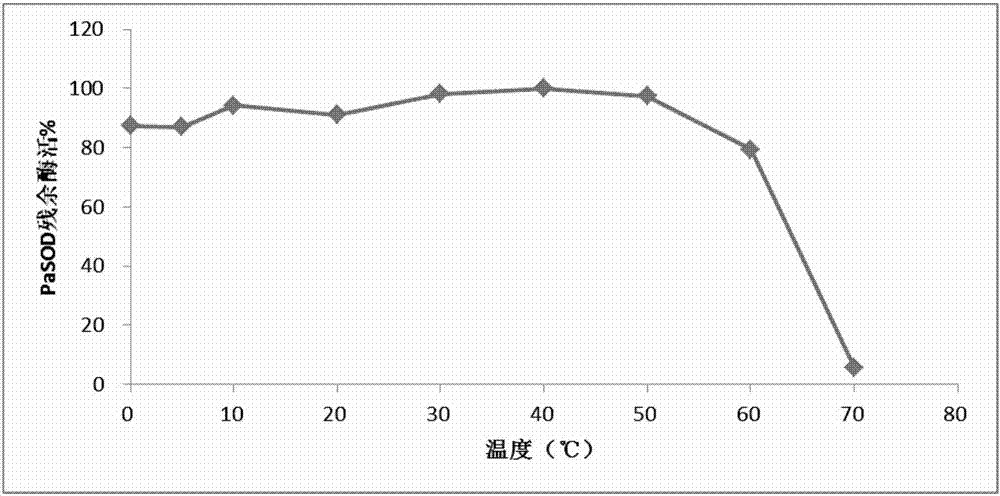
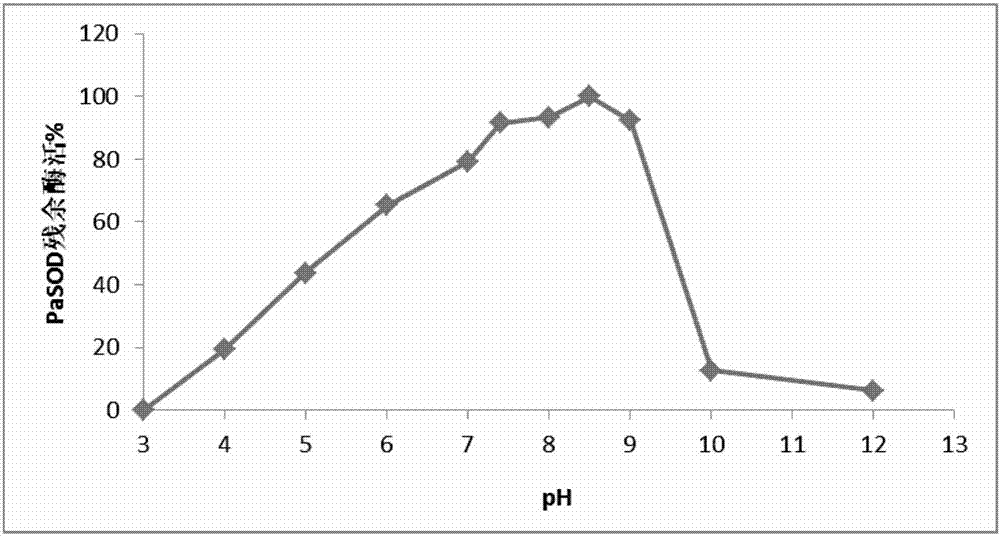
[0041] Example 3 Soluble expression and protein purification of Paelopatides sp. superoxide dismutase PaSOD gene:
[0042] The correctly sequenced DH5α was expanded and cultivated, and the plasmid was extracted. PaSOD carried out Bam H I and Pst I double enzyme digestion reactions on the plasmid and the pCold II vector plasmid at the same time. The PaSOD target gene and the linear pCold II vector were recovered from the gel, and the two were ligated overnight with TAKARA T4 ligase (TAKARA company), and the ligated product was transformed into Escherichia coli DH5α competent cells, and a single colony was picked for PCR and then the target DNA fragment was selected. Positive clones were sequenced.
[0043] The correctly sequenced DH5α was expanded and cultured, the plasmid was extracted, and transformed into Escherichia coli Chaperone CompetentCell pG-KJE8 / BL21. After spreading the plate, a single colony was picked for PCR to verify the correctness of the transformation. Sele...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com