Alginate lyase SHA-6 gene and application thereof
A technology of alginate lyase and SHA-6, applied in the field of microbial genetic engineering, can solve the problems of unreported and low expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1: Marina catena alginatilytica Preparation and Detection of SH-52 Genomic DNA
[0040] used in the present invention Marina catena alginatilytica SH-52 is a strain screened by our laboratory. The preparation of SH-52 genomic DNA adopts the extraction method of common bacterial genome. The specific content is as follows: Take 2 mL of overnight culture liquid and centrifuge at 4000 rpm for 2 min at 4 ° C. Discard the supernatant and collect the bacteria. To the body, add 100 µL Solution I suspension, 30 µL 10% SDS and 1 µL 20 mg / mL proteinase K, mix well, and incubate at 37°C for 1 h. Add 100 µL of 5M NaCl, invert and mix well, then add 20 µL CTAB / NaCl solution (CTAB 10%, 0.7M NaCl), and incubate at 65°C for 10 min. Add an equal volume of chloroform / isoamyl alcohol (24:1), mix by inverting, and centrifuge at 12,000 rpm for 5 min at 4°C. Take the supernatant, add 2 times the volume of absolute ethanol, 0.1 times the volume of 3M NaAc, place at -20°C for 30 m...
Embodiment 2
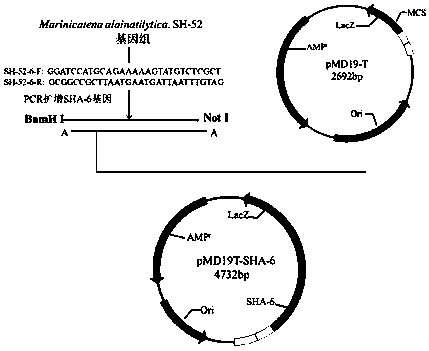
[0041] Embodiment 2: the amplification and TA clone of alginate lyase SHA-6 gene
[0042] alginate lyase SHA-6 gene amplification and cloning figure 2 As shown, design a pair of specific primers, the sequence is as follows:
[0043] SHA-6-F : GGATCC ATGCAGAAAAAGTATGTCTCGCT
[0044] SHA-6-R: GCGGCCGC TTAATGAATGATTAATTTGTAG
[0045] The GGATCC characteristic sequence was introduced at the 5' end to form a BamH I restriction site; the GCGGCC characteristic sequence was introduced at the 3' end to form a NotI restriction site.
[0046] Add 10 ng of Marinicatena alginatilytica SH-52 genomic DNA was used as a template, and 50ng of specific primers SHA-6-F and SHA-6-R, 2.5µL of dNTP (10mM), 2.5µL of Pfu reaction buffer and 0.5µL of Pfu (5U / µL) were added to aggregate Enzyme (Beijing Quanshijin Biotechnology Co., Ltd.), add double distilled water to make the final volume 25µL. Heat at 94°C for 3 minutes on the PCR instrument, then perform 25 cycles of reaction according t...
Embodiment 3
[0047] Example 3: Prokaryotic expression vector pET-32a- SHA-6 build
[0048] Such as Figure 4 As shown, the purified prokaryotic expression vectors pET-32a and pMD19-T- SHA-6 , the cut vector and the inserted target fragment were separated by agarose gel electrophoresis, and the vector fragments pET-32a and pMD19-T- SHA-6 produced by cutting SHA-6 The DNA fragment was connected to the pET-32a vector fragment and SHA-6 Prokaryotic expression vector pET-32a- SHA-6 . Transform the ligation reaction mixture into Escherichia coli competent cells BL21 (Tiangen Biochemical Technology), spread the transformed Escherichia coli on a plate added with ampicillin (final concentration: 100 µg / mL), culture overnight at 37°C, and select Amp Resistant recombinant colonies, using SHA-6-F and SHA-6-R primers for colony PCR verification, after the successful connection of the plasmids for liquid culture, the plasmid DNA was extracted by alkaline lysis, and the recombinant plasmids ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com