Recombinant escherichia coli expressing formamidase and phosphite dehydrogenase fusion protein, construction method and application thereof
A technology of phosphite dehydrogenase and recombinant Escherichia coli, which is applied in the field of genetic engineering and can solve problems such as common bacteria
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
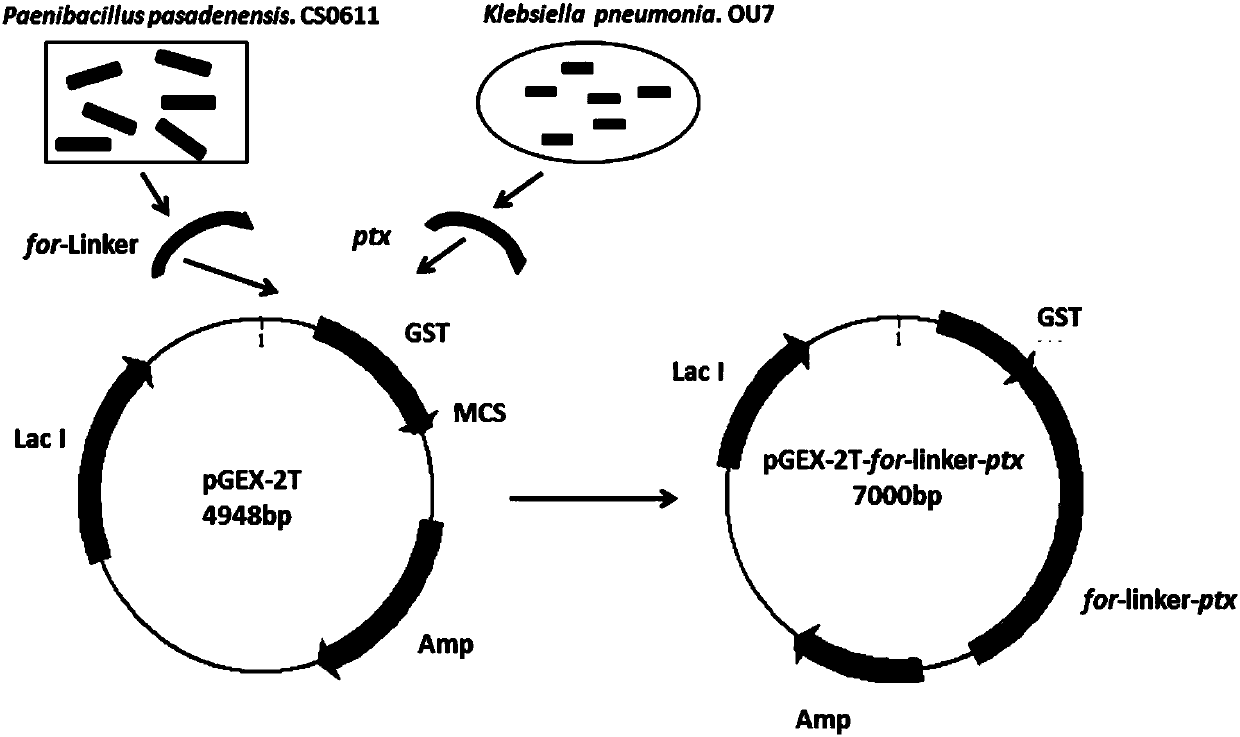
[0051] Acquisition of formamidase gene for (including Linker sequence)
[0052] Paenibacillus pasadenensis.CS0611 was cultured in LB medium at 37°C and 180rpm for one day; the cultured cells were collected at 4°C, 8000rpm and centrifuged for 5min, and washed twice with normal saline to remove residual culture The genome of Paenibacillus pasadenensis.CS0611 was extracted according to the specific method of the OMEGA bacterial genome DNA extraction kit;
[0053] Using the extracted Paenibacillus pasadenensis.CS0611 genome as a template, and A2(5'- CGACCCACCA CCGCCCGAGCCACCGCCACC TCGCGCCGCGCCTCCCTTCG C-3') are the upstream and downstream primers respectively, and the formamidase gene for-Linker is amplified by PCR;
[0054] The enzyme reagents used in PCR are TaKaRa company HS DNA Polymerase with GCbuffer; PCR reaction system and conditions are as follows:
[0055] Composition of PCR reaction solution (25μL)
[0056]
[0057] PCR reaction conditions: 94°C for 5 min...
Embodiment 2
[0060] The acquisition of phosphite dehydrogenase gene ptx
[0061] The phosphite dehydrogenase gene is derived from the self-screened Klebsiella pneumonia.OU7, obtained through self-screening;
[0062] The self-screened Klebsiella pneumonia.OU7 genome was extracted according to the specific method of the OMEGA bacterial genome DNA extraction kit, and the self-screened Klebsiella pneumonia. - TGGCTCGGGCGGTGGTGGGTCGATGCTGCCGAAACTCGTTATA-3') and The upstream and downstream primers are respectively used to amplify the phosphite dehydrogenase gene ptx by PCR;
[0063] The enzyme reagents used in PCR are TaKaRa company HS DNA Polymerase with GCbuffer; PCR reaction system and conditions are as follows:
[0064] Composition of PCR reaction solution (25μL)
[0065]
[0066]
[0067] PCR reaction conditions: 94 °C for 5 min; then 98 °C for 10 s, 55 °C for 5 s, 72 °C for 70 s, and cycle 30 times; then 72 °C for 7 min; and finally cooling to 16 °C.
[0068] The DNA product...
Embodiment 3
[0070] The fusion gene for-Linker-ptx was obtained by overlapping PCR amplification
[0071] Take the amplified for-Linker and ptx fragments as templates, and use primers and primers The upstream and downstream primers, respectively, were amplified to obtain the fusion gene for-Linker-ptx.
[0072] The amplification conditions were as follows: 94°C for 5 min; then 98°C for 10 s, 55°C for 5 s, 72°C for 150 s, and cycled 30 times; finally, 72°C for 7 min; and finally cooling to 16°C.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com