Method of knocking out T cell immunity detecting point pathway genes and application
A gene knockout and cellular immunity technology, applied in the field of genetic engineering, can solve the problems of expensive antibody drugs, imbalanced immune tolerance, and no immune response at immune checkpoints, and achieves high safety and a simple and easy method. , effective effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0065] Example 1 sgRNA design and construction
[0066] (1) sgRNA primer pair design
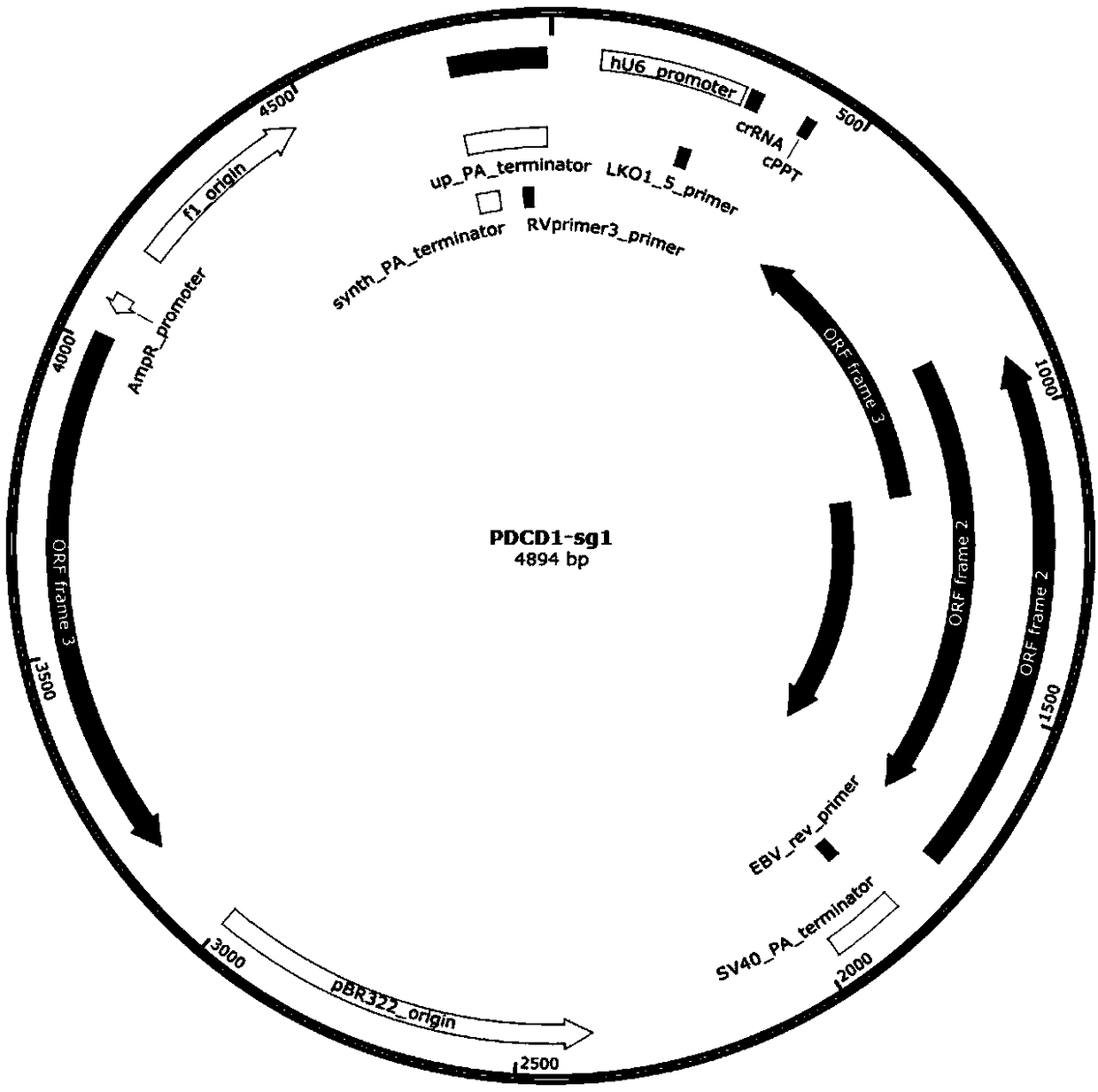
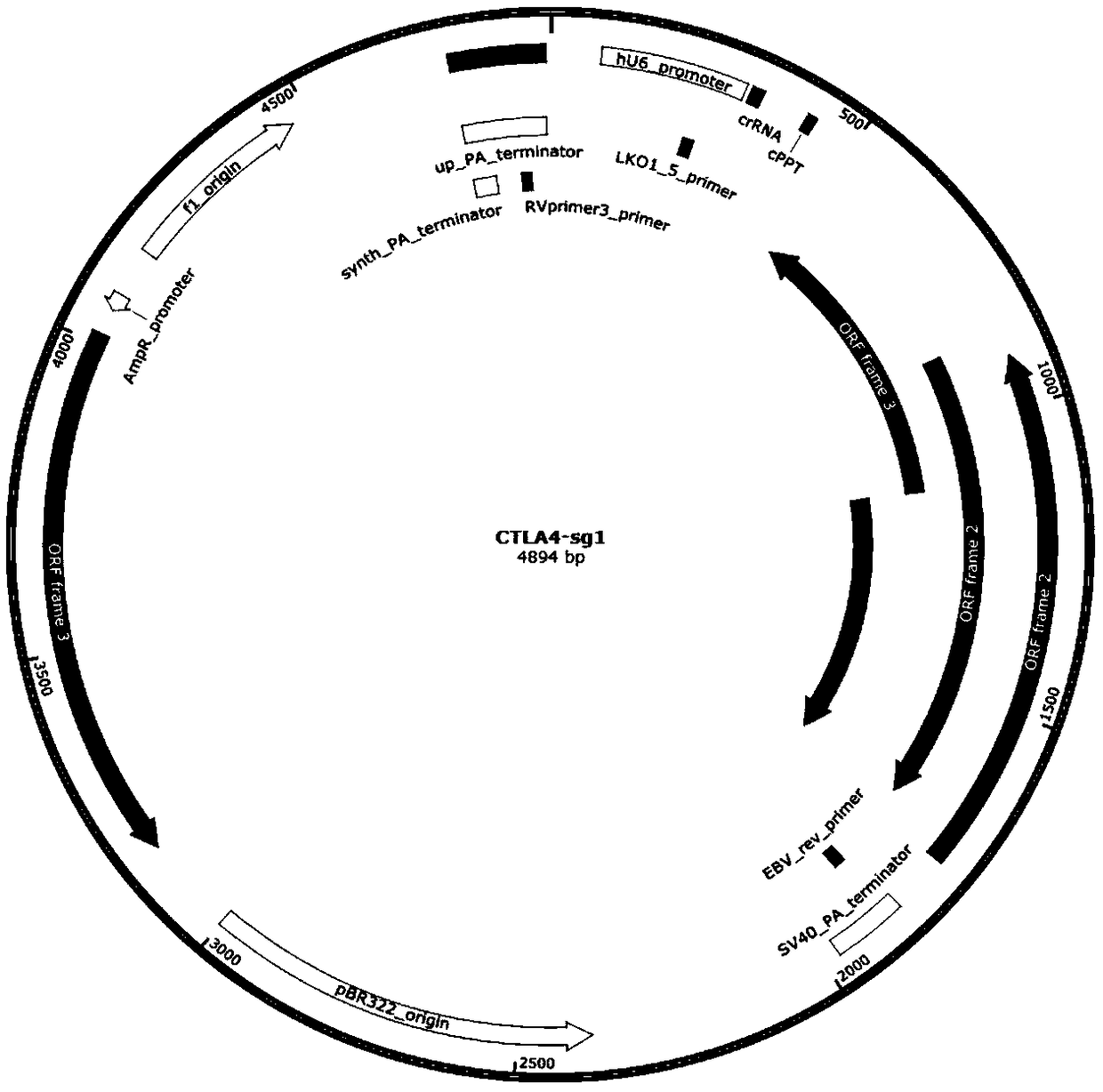
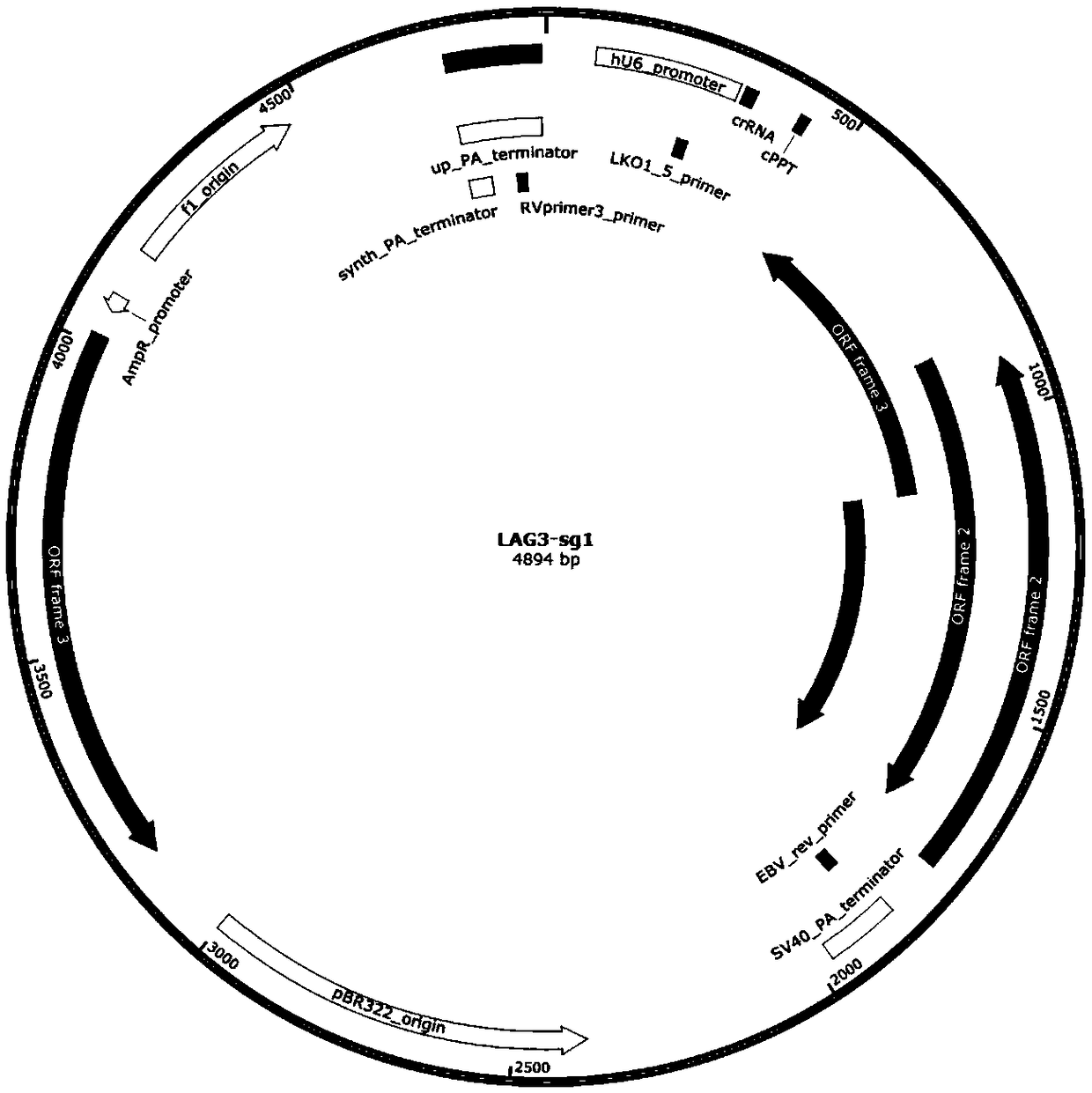
[0067] Select the target DNA region of the T cell immune checkpoint pathway gene. The T cell immune checkpoint pathway genes are: cytotoxic T lymphocyte-associated protein (T-lymphocyte antigen-4, CTLA-4), programmed death receptor-1 (programmed death-1, PD-1), lymphocyte activation gene-3 (Lymphocyte activation gene-3, LAG-3), T-cell immunoglobulin and mucin-3 (T-cell immunoglobulin and mucin-3, TIM- 3), according to the selected target sequence, a pair of sgRNA was synthesized for each target gene, the sequence is as follows:
[0068]
[0069]
[0070] (2) Denature and anneal each pair of sgRNAs in the above table
[0071] The annealing system is:
[0072]
[0073] Run in the PCR machine according to the following landing PCR program: 95°C, 5min; 95-85°C at-2°C / s; 85-25°C at-0.1°C / s; 4°C, save.
[0074] (3) Linearization of pGL3-U6-sgRNA plasmid
Embodiment 2
[0085] Example 2 Functional verification of sgRNA
[0086] (1) Cell culture and transfection
[0087] 1) HEK293T cells were inoculated and cultured in DMEM high-glucose medium (HyClone, SH30022.01B), which contained 10% FBS, penicillin (penicillin, 100 U / ml) and streptomycin (streptomycin, 100 μg / ml);
[0088] 2) Inoculate HEK293T cells in a 12-well plate before transfection, and when the cells grow to 70%-80% density of the 12-well plate, replace with antibiotic-free medium to prepare for transfection;
[0089] 3) According to Lipofectamine TM 2000 Transfection Reagent (Invitrogen, 11668-019) operating manual, mix 0.5 μg pGL3-U6-hPD1sg1 and 1.5 μg pST1374-h13Lb-cpf1 plasmid, co-transfect into cells in each well, change the medium after 6 hours, Add blasticidin (Blasticidin, invitrigen, ant-bl-1) and puromycin (Puromycin, invitrigen, ant-pr-1) drug sieve, and collect the cells after 48 hours.
[0090] (2) T7EN1 enzyme digestion detection
[0091] 1) Extract DNA from the c...
Embodiment 3
[0098] Example 3 Preparation of PBMC cells
[0099] Use an anticoagulant tube to collect peripheral blood from healthy people, and shake it while collecting to fully mix the peripheral blood with the anticoagulant; slowly add the anticoagulant blood to a 50ml centrifuge tube filled with an equal volume of lymphocyte separation medium (Ficoll), 450g, Slowly ascend and descend slowly for 25 minutes, and the centrifugation cannot be stopped in the middle. After the centrifugation, carefully absorb the buffy coat cells above the lymphocyte separation solution, transfer to a new 50ml centrifuge tube, add PBS, 300g, and slowly ascend and descend for 10 minutes. , discard the supernatant, and keep the cell pellet at the bottom of the centrifuge tube; add PBS again, 160g, centrifuge slowly for 15min, discard the supernatant; finally add PBS, 300g, centrifuge slowly for 10min, discard the supernatant, and obtain PBMC cell.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com