Sl1ARF10 gene and Sl1miR160b regulating tomato lateral bud development and application thereof
A technology of slarf10-srdx and lateral buds, applied in the fields of application, genetic engineering, plant genetic improvement, etc., can solve problems such as undiscovered effects, and achieve the effect of increasing the incidence
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Embodiment 1 Tomato SIARF10 promoter cloning and GUS staining analysis
[0039] (1) Sequence analysis and cloning of SIARF10 promoter
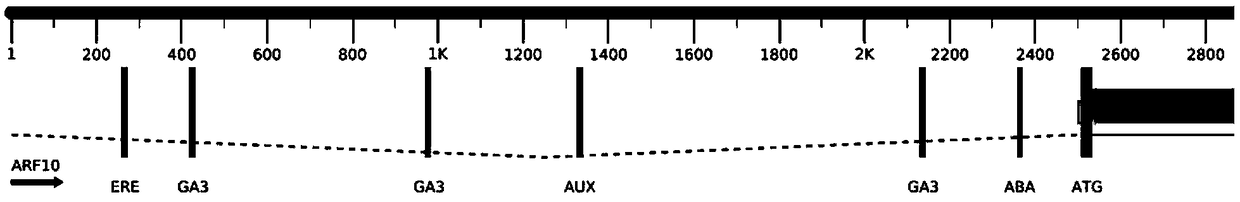
[0040] Using PlantCARE (http: / / bioinformatics.psb.ugent.be / webtools / plantcare / html / ) online software to analyze the sequence of about 2.5kb of the upstream promoter pSlARF10 of the tomato SlARF10 gene, the pSlARF10 promoter contains a variety of hormone-regulated response elements, Including auxin response element AUX, gibberellin response element GA3, ethylene response element ERE, etc., the results are as follows figure 1 shown.
[0041] According to the nucleotide sequence of pSIARF10 (SEQ ID NO.4), design specific primers pSIARF10-F and pSIARF10-R, and use CTAB method (RTG2405-01, Zhongke Tairui) to extract tomato genomic DNA as a template for conventional PCR amplification target fragment.
[0042] The primer sequences are as follows:
[0043] pSlARF10-F: 5'-CAATTTACATATACAACACATGCCTCA-3'
[0044] pSlARF10-R: 5'-GCTAATCCAATAGT...
Embodiment 2
[0067] Example 2 Construction of recombinant expression vector P35S-SlmiR160b-NOS and genetic transformation of tomato
[0068] (1) Cloning of tomato SlmiR160b
[0069] Through bioinformatics analysis, it was found that in the tomato genome, in addition to the published SlmiR160a Genbank (NR_107984.1) or miRBase (MI0008357), miR160 also has another site SlmiR160b on chromosome 12 (GenBank: HG975524.1) (SEQ ID NO. 1). Utilize The mfold Web Server online service software RNA Folding Form (version 2.3energies) to analyze the SlmiR160b hairpin structure such as Figure 4 shown.
[0070] According to the precursor nucleotide sequence of SlmiR160b (SEQ ID NO.1), specific primers SlmiR160b-F and SlmiR160b-R were designed, and the tomato genomic DNA was extracted by CTAB method (RTG2405-01, Zhongke Tairui) as a template for routine PCR amplification of target fragments. The primer sequence is:
[0071] SlmiR160b-F: 5'-CCAAGTCCAAGAAATGAAATTG-3'
[0072] SlmiR160b-R: 5'-GTTGCCAATTGG...
Embodiment 3
[0102] Example 3 Construction of recombinant expression vector P35S-SIARF10-RNAi-NOS and genetic transformation of tomato
[0103] (1) Cloning of tomato SIARF10-RNAi target fragment
[0104] Take Micro Tom tomato plants in flowering stage as material, use TRIzol TM Plus RNA Purification Kit (12183555, Invitrogen TM ), extract tomato total RNA according to the instructions, and use DNase I (18047019, Invitrogen TM ) to remove residual traces of DNA, and use a spectrophotometer to measure the concentration of RNA for later use.
[0105] About 2.0 μg of tomato total RNA was taken, and the first-strand cDNA was synthesized using PrimeScript II first-strand cDNA synthesis kit (6210A, Takara) according to the instructions.
[0106] According to the tomato SIARF10 gene sequence (SEQ ID NO.2), a specific segment was found by comparing homologous gene sequences, and specific primers SIARF10-RNAi-F and SIARF10-RNAi-R were designed. Using cDNA as a template, the specific fragment of...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com