Construction method of gene detection library of hereditary hypertrophic cardiomyopathy and kit
A technology for hypertrophic cardiomyopathy and gene detection, which is applied in chemical libraries, biochemical equipment and methods, and microbial determination/inspection.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
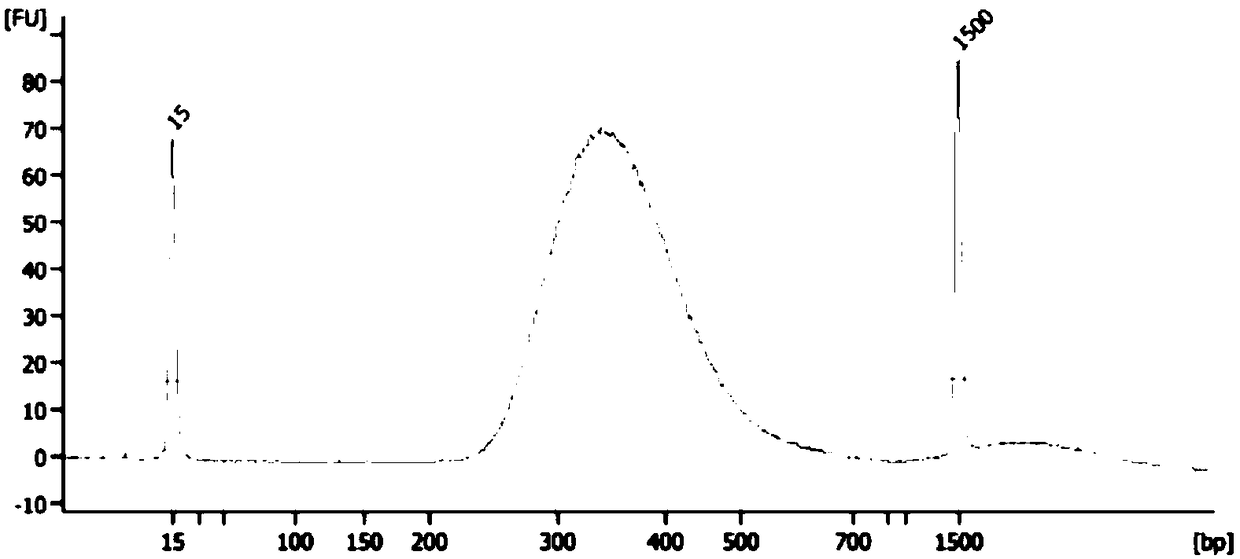
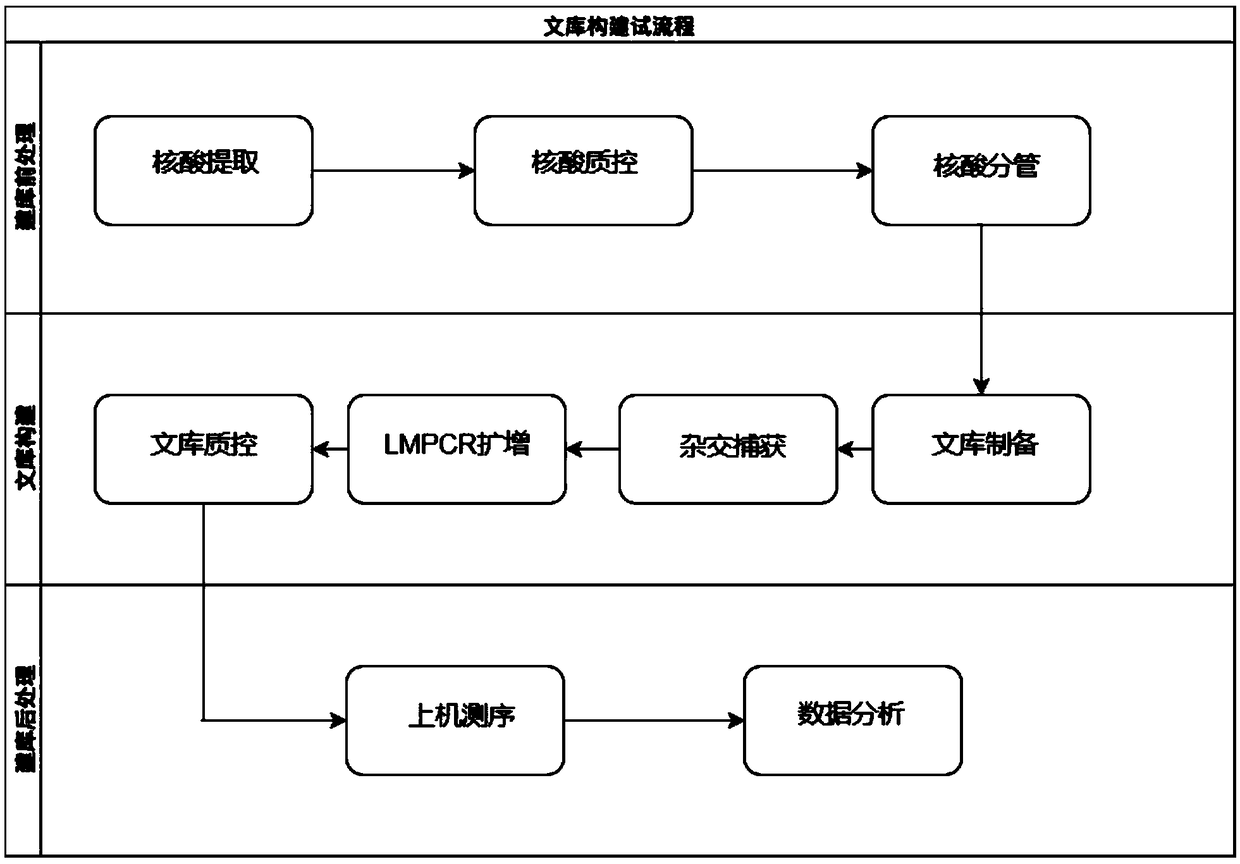
[0103] Four peripheral blood samples were used for library construction, using the full coding region and variable splicing region (exon to intron extension) of ACTC1, ACTN2, MYBPC3, MYH7, MYL2, MYL3, TNNI3, TNNT2, TPM1 genes 20bp) as the primer pool of the target region for multiplex reaction PCR, combined with the high-throughput sequencing platform Miseq for DNA sequencing, and then detected point mutations (SNP) and small fragment insertion-deletion (InDel). The specific operation process is as follows:
[0104] (1) Nucleic acid extraction and quality inspection: After nucleic acid extraction and quality inspection of peripheral blood samples, it is required to meet certain quality control standards: DNA integrity is good, no protein RNA pollution, DNA concentration ≥ 20ng / μL; DNA purity: OD260 / 280 1.8-2.0, OD260 / 230>2.0; total DNA input amount: 0.1μg-1μg.
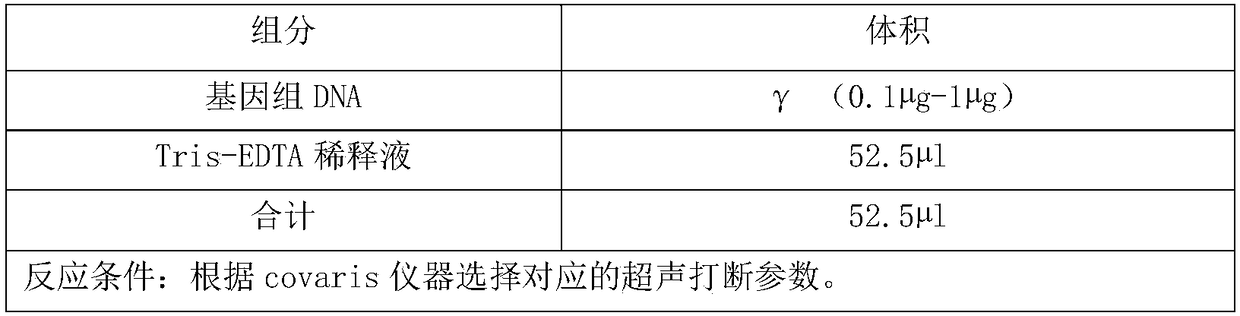
[0105](2) Library preparation: Using the above DNA, make up to 52.5 μl with TRIS-EDTA after sampling, perform ultr...
Embodiment 2
[0113] 16 buccal swab samples were used for library construction, using the full coding region and variable splicing region (exon to intron extension) of ACTC1, ACTN2, MYBPC3, MYH7, MYL2, MYL3, TNNI3, TNNT2, TPM1 genes 20bp) as the primer pool of the target region for multiplex reaction PCR, combined with the high-throughput sequencing platform Nextseq550AR for DNA sequencing, and then detecting point mutations (SNPs) and small fragment insertions and deletions (InDels). The specific operation process is as follows:
[0114] (1) Nucleic acid extraction and quality inspection: After nucleic acid extraction and quality inspection of peripheral blood samples, it is required to meet certain quality control standards: DNA integrity is good, no protein RNA pollution, DNA concentration ≥ 20ng / μL; DNA purity: OD260 / 280 1.8-2.0, OD260 / 230>2.0; total DNA input amount: 0.1μg-1μg.
[0115] (2) Library preparation: Using the above DNA, make up to 52.5 μl with TRIS-EDTA after sampling, pe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com