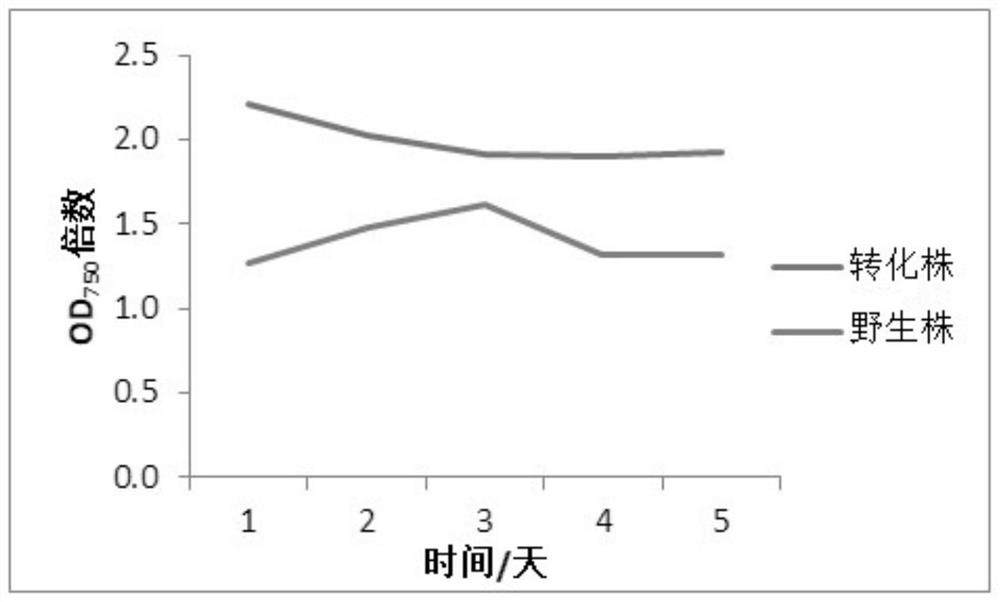
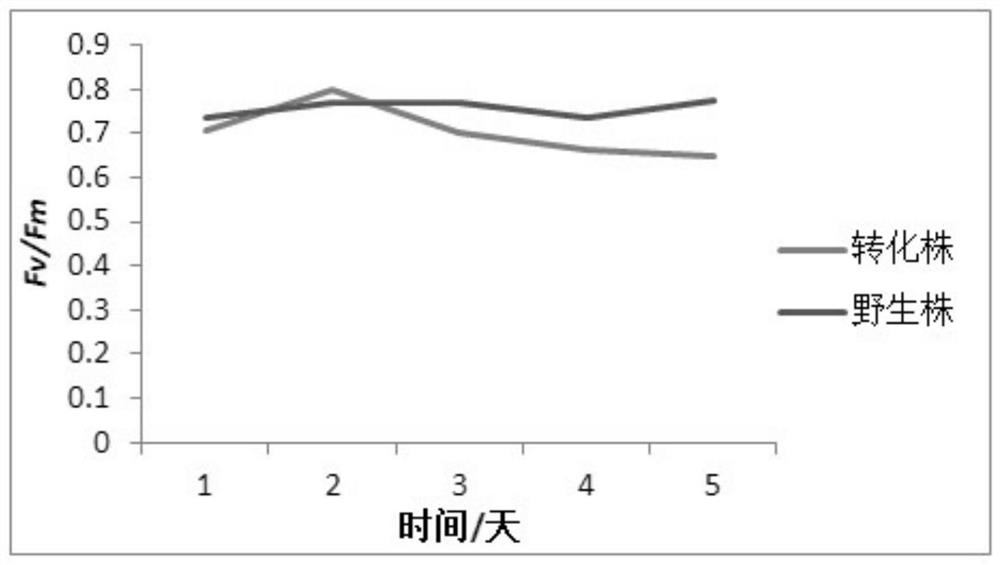
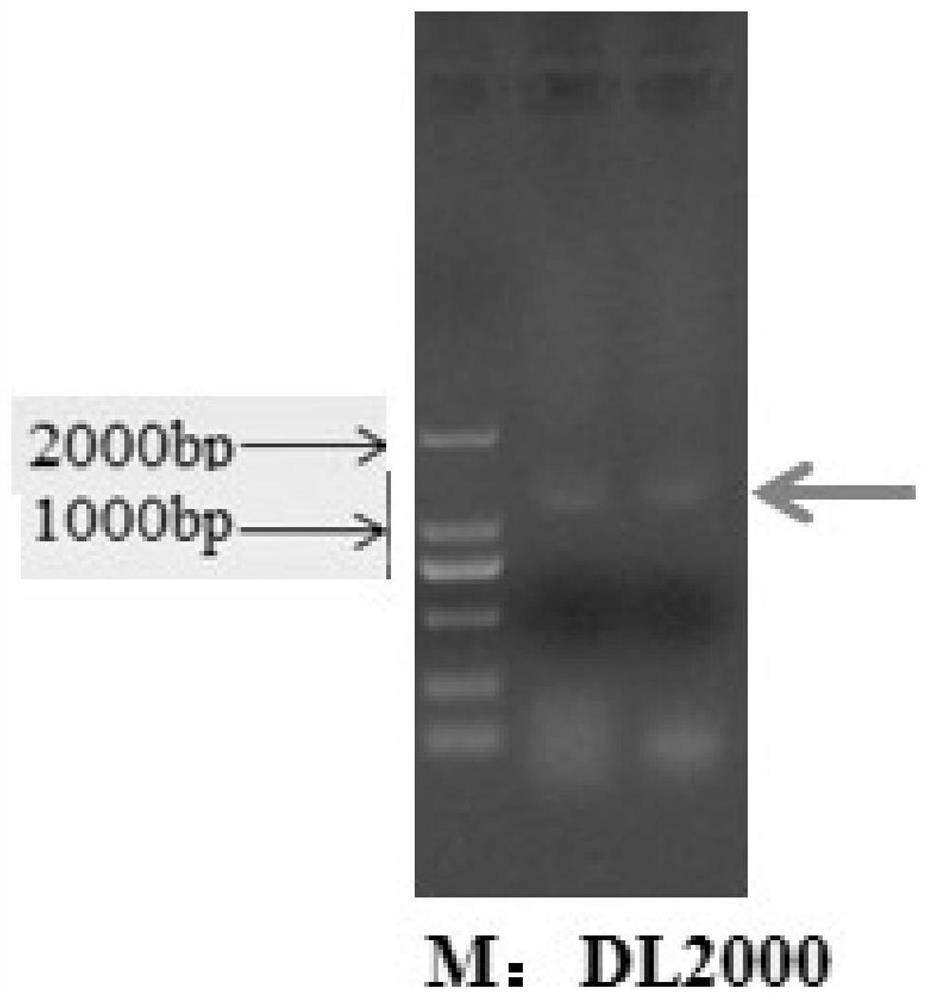
A method for improving carbon fixation efficiency of microalgae and transgenic Chlamydomonas and application
A transgenic, high-efficiency technology, applied in the field of biological genetic engineering, to achieve the effects of increasing growth rate, accelerating growth rate, and improving carbon sequestration efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] Example 1: Transformation of the foreign gene corresponding to the GAP3 gene in Chlamydomonas reinhardtii and construction of the expression vector
[0059] The algal strain selected in this example is Chlamydomonas reinhardtii cc-137 (purchased from the American Chlamydomonas Center) as the recipient of the transgenic operation, and the algal strain is a wild-type Chlamydomonas reinhardtii strain.
[0060] The medium used in the cultivation of Chlamydomonas reinhardtii is TAP medium, and the formula of TAP medium is as follows:
[0061] Mother liquor 1 (salt solution): 20gNH 4 Cl, 5g MgSO 4 ·7H 2 O, 2.5g CaCl 2 2H 2 O is fixed in 500ml deionized water;
[0062] Mother liquor 2 (phosphate solution): 10.8gK 2 HPO 4 , 5.6gKH 2 PO 4 Set the volume to 500ml deionized water;
[0063] Mother liquor 3 (Hutner's trace metal salt solution): 50.0gNa 2 -EDTA·2H 2 O,22gZnSO 4 ·7H 2 O, 5.06gMnCl 2 4H 2 O, 1.61g CoCl 2 ·6H 2 O, 1.57g CuSO 4 ·5H 2O, 1.10g (NH 4 ) ...
Embodiment 2
[0074] Example 2: Genetic transformation of Chlamydomonas reinhardtii
[0075] The recombinant plasmid pChlamy-c-GAP3 was extracted by conventional SDS alkaline lysis method
[0076] The specific steps of the "electrotransfer method" are as follows:
[0077] (1) Cultivate wild-type Chlamydomonas reinhardtii cc-137 (Chlamydomonas Resource Center CC-137) in continuous light and TAP medium to OD=0.4-0.6, and dilute the cells to OD=0.05-0.08. After 24 hours of culture, algae When the liquid concentration reaches 0.3-0.6, take 15ml of algae cells at 2500rpm, centrifuge at room temperature for 10min, and discard the supernatant.
[0078] (2) Resuspend the algae cells with 250 μl of 40 mM sucrose TAP solution, add to the electric shock cup, and then add 1 to 2 μg of linearized pChlamy-c-GAP3 plasmid to the cup for electric shock (600V, 50uF, infinite resistance).
[0079] (3) After the electric shock, the mixture of algal cells and exogenous DNA in the electric shock cup was added ...
Embodiment 3
[0082] Example 3: Screening and identification of transgenic Chlamydomonas reinhardtii
[0083] The expression vector of C. reinhardtii contains Hygromycin resistance gene, so it has hygromycin resistance, and the successfully transformed algal strains can grow on the plate containing hygromycin resistance. The detection of transgenic algae includes PCR verification of single algae colony at the gene level and western blotting verification at the protein level.
[0084] (1) Double PCR verification of the single algal fall of the transformed strain at the gene level
[0085] Pick the single algae on the plate and drop it into the PCR tube, add 10 μl of sterile water to resuspend the algae cells, cook at 98°C for 20-30 minutes, centrifuge to get the supernatant, use PC-F / R primers, (Use PrimeSTAR Max DNA Polymerase , TaKaRa, Dalian City, CodeNo.R045Q), using PCR technology to amplify the target gene fragment from the algae, and then using the GAP3-specific primer (GAP3-F / R) on ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com