LAMP primer combination for detecting candida albicans in intraocular fluid and application thereof
A Candida albicans, primer combination technology, applied in microorganism-based methods, microorganisms, recombinant DNA technology, etc., can solve the problems of easy contamination, high false positive rate, application limitations, etc., and achieve high specificity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Embodiment 1, primer design and preparation
[0058] A large number of sequence analyzes and comparisons were carried out to obtain several primers for identifying Candida albicans. Preliminary experiments were performed on each primer to compare performances such as sensitivity and specificity, and finally a LAMP primer set for identifying Candida albicans was obtained.
[0059] The primer set used to identify Candida albicans, including 2 outer primers (F3, B3), 2 inner primers (FIP, BIP) and 2 loop primers (LF, LB), each primer sequence is as follows (5 '→3'):
[0060] F3 (sequence 1 of the sequence listing): CgATACgTAATATgAATTgCAgAT;
[0061] B3 (sequence 2 of the sequence listing): CCATTgTCAAAgCgATCC;
[0062] FIP (SEQ ID NO: 3 of the Sequence Listing): AggCATgCCCTCCggAATACTCgTgAATCATCgAATCTTTgAAC;
[0063] BIP (SEQ ID NO: 4 of the Sequence Listing): gAgCgTCgTTTCTCCCTCAAACCgCCTTACCACTACCgTCTT;
[0064] LF (Sequence 5 of the Sequence Listing): AgAgggCgCAATgTgC;...
Embodiment 2
[0066] Embodiment 2, detection method establishment
[0067] 1. Extract the genomic DNA of the fungus to be tested or extract the total DNA of the biological sample to be tested.
[0068] 2. Take the DNA obtained in step 1 as a template, and use the LAMP primer set prepared in Example 1 to perform LAMP amplification.
[0069] Reaction system for LAMP amplification: 1μL 10×ThermoPol Buffer, 1.6μL 5M betaine, 0.1μL 50mg / ml BSA, 0.4μL 100mM MgSO 4 , 0.3 μL 20×EvaGreen, 0.15 μL 100 mM dNTPs, 0.4 μL 8U / ml Bst DNA polymerase large fragment, 1 μL primer mix (mixture of F3, B3, FIP, BIP, LF, and LB), 2ng template DNA, with ddH 2 O to make up to 10 μL. The concentration of each primer in the reaction system is as follows: 0.5 μM F3, 0.5 μM B3, 2 μM FIP, 2 μM BIP, 1 μM LF, 1 μM LB.
[0070] Reaction program for LAMP amplification: constant temperature at 65°C for 50min. During the reaction process, the fluorescent signal was detected by a fluorescent PCR instrument.
[0071] If a p...
Embodiment 3
[0072] Embodiment 3, specificity
[0073] Bacteria to be tested: Candida albicans, Aspergillus fumigatus, Aspergillus flavus and Candida glabrata.
[0074] The genomic DNA of the bacteria to be tested was extracted, and the detection method in Example 2 was used for detection. Set up a negative control using water as template DNA.
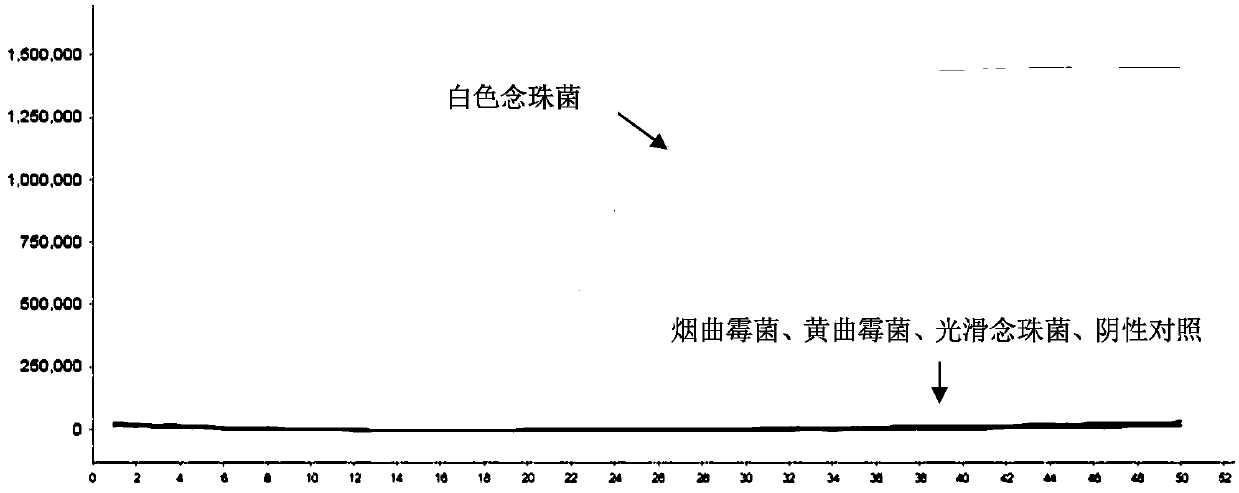
[0075] see results figure 1 . figure 1 In , the abscissa is the cycle number, and the ordinate is the fluorescence signal intensity. The results showed that Candida albicans genomic DNA obtained S-type amplification curve, and the amplification result was positive, while Aspergillus fumigatus genomic DNA, Aspergillus flavus genomic DNA and Candida glabrata genomic DNA did not obtain S-type amplification curve, and the amplification result was positive. is negative.
[0076] The results show that the LAMP primer set prepared in Example 1 has high specificity.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com