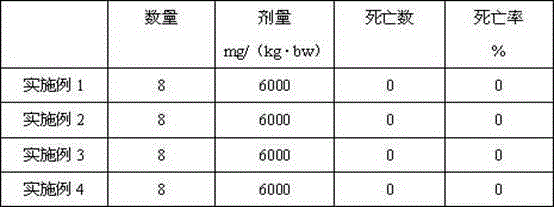
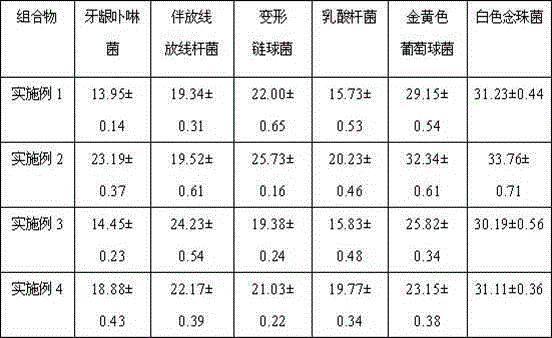
Patents
Literature
684 results about "Monilinia fructicola" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Monilinia fructicola is a species of fungus in the order Helotiales. A plant pathogen, it is the causal agent of brown rot of stone fruits.
PCR identification and quantification of important Candida species
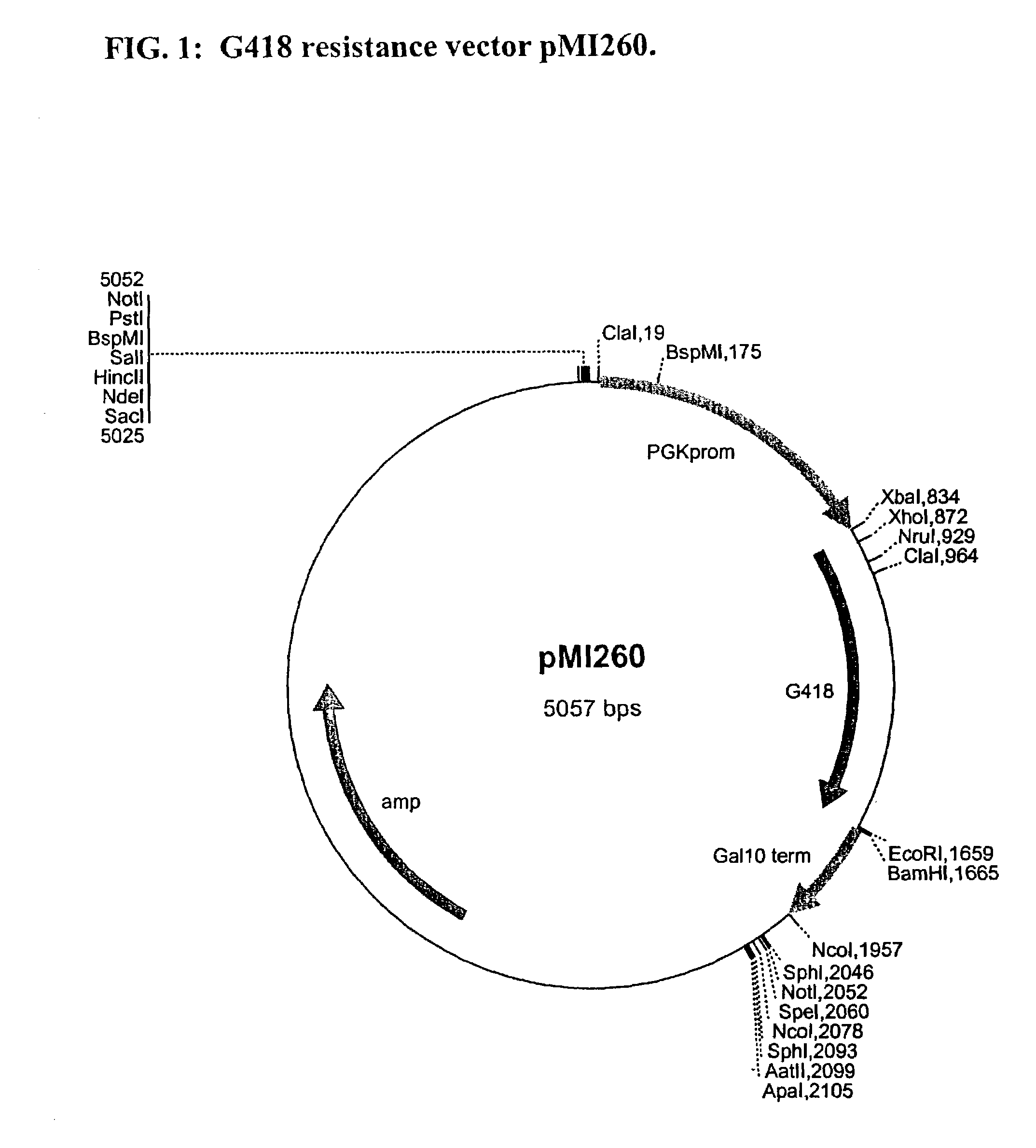
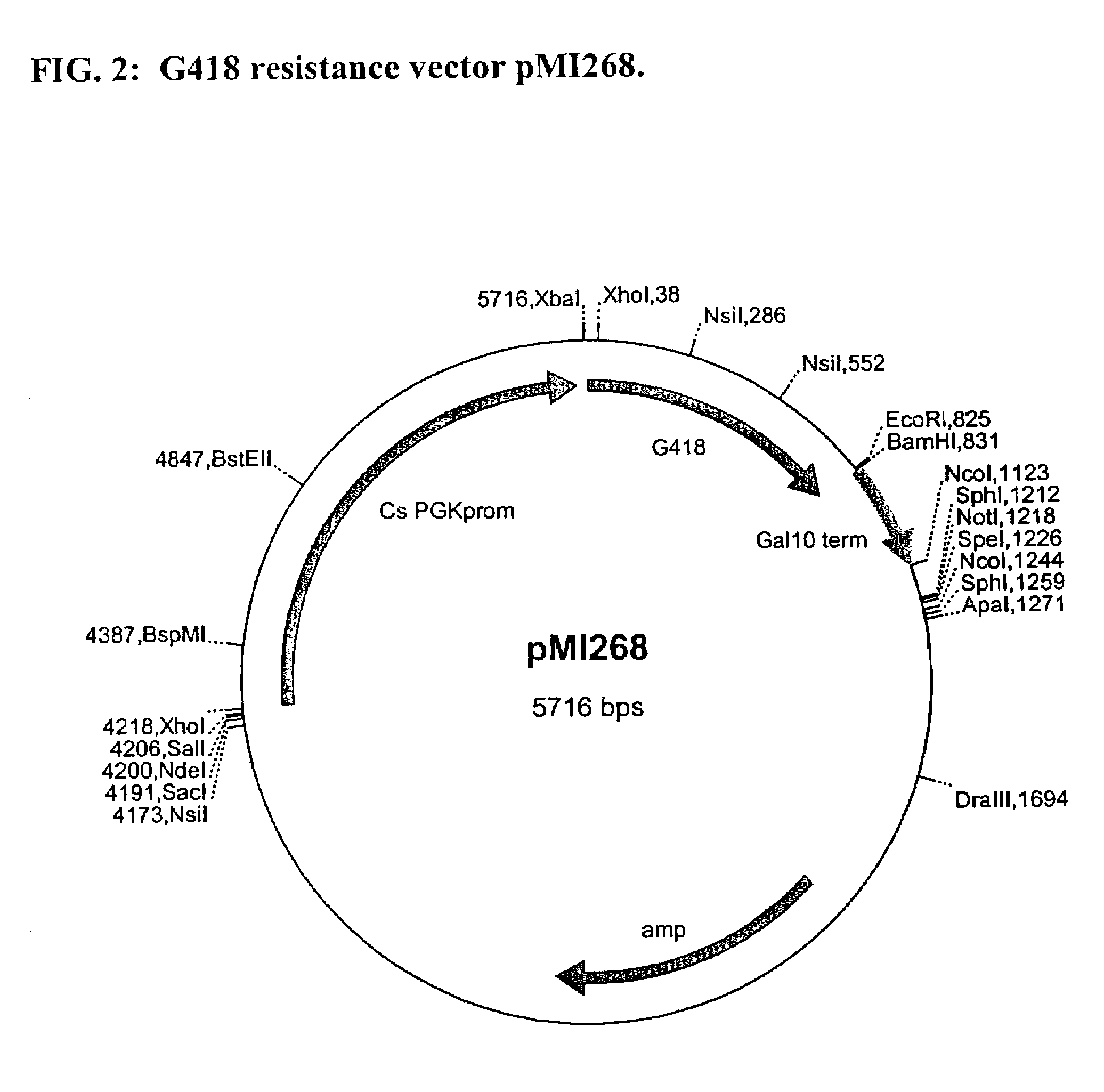
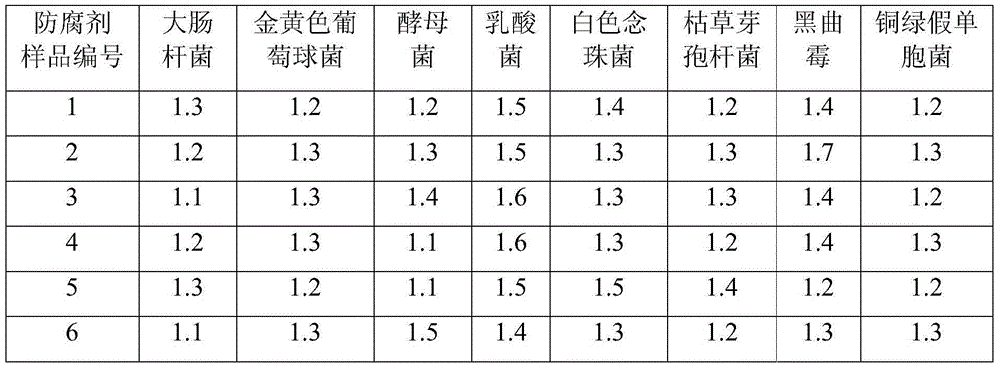
InactiveUS6017699AConvenient amountHigh sensitivitySugar derivativesMicrobiological testing/measurementQuantitative determinationCombined use
The subject invention relates to a set of DNA primers which, when utilized in conjunction with the polymerase chain reaction (PCR) assay, can amplify and speciate DNA from five medically important Candida species. Furthermore, the PCR amplified products, generated by the primers, can also be used to create species specific probes which can also detect and confirm the five species of Candida. Thus, the present invention allows for early diagnosis and treatment of an infection. The assay is useful in the context of monitoring antifungal treatment regimens, screening potential antifungal agents, and similar applications requiring quantitative determinations.
Owner:UNIV OF PITTSBURGH THE
Pharmaceutical compositions and methods to vaccinate against candidiasis
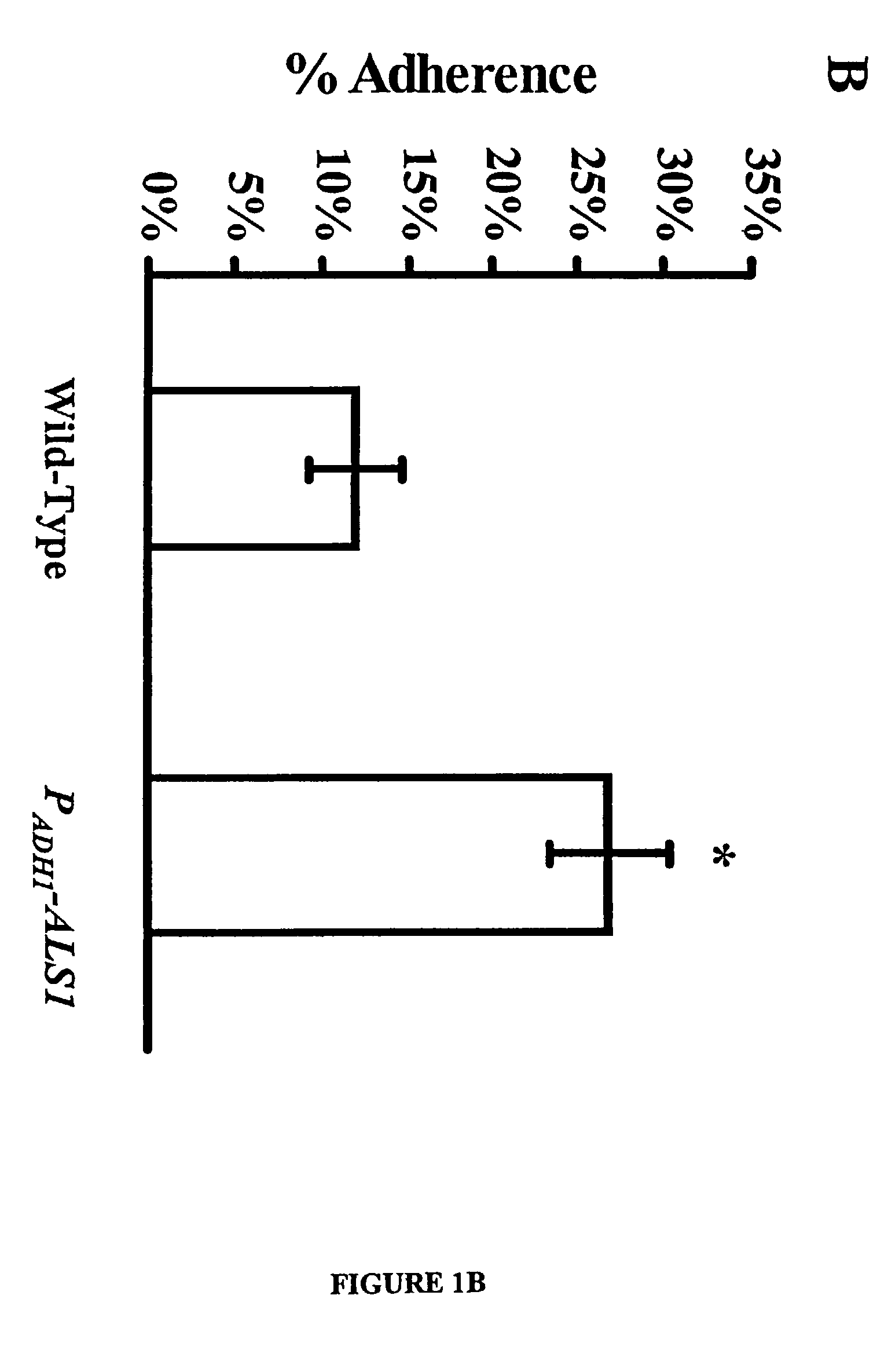
InactiveUS20030124134A1Treat prevent alleviateToxic reductionPeptide/protein ingredientsSnake antigen ingredientsHospitalized patientsVirulent characteristics
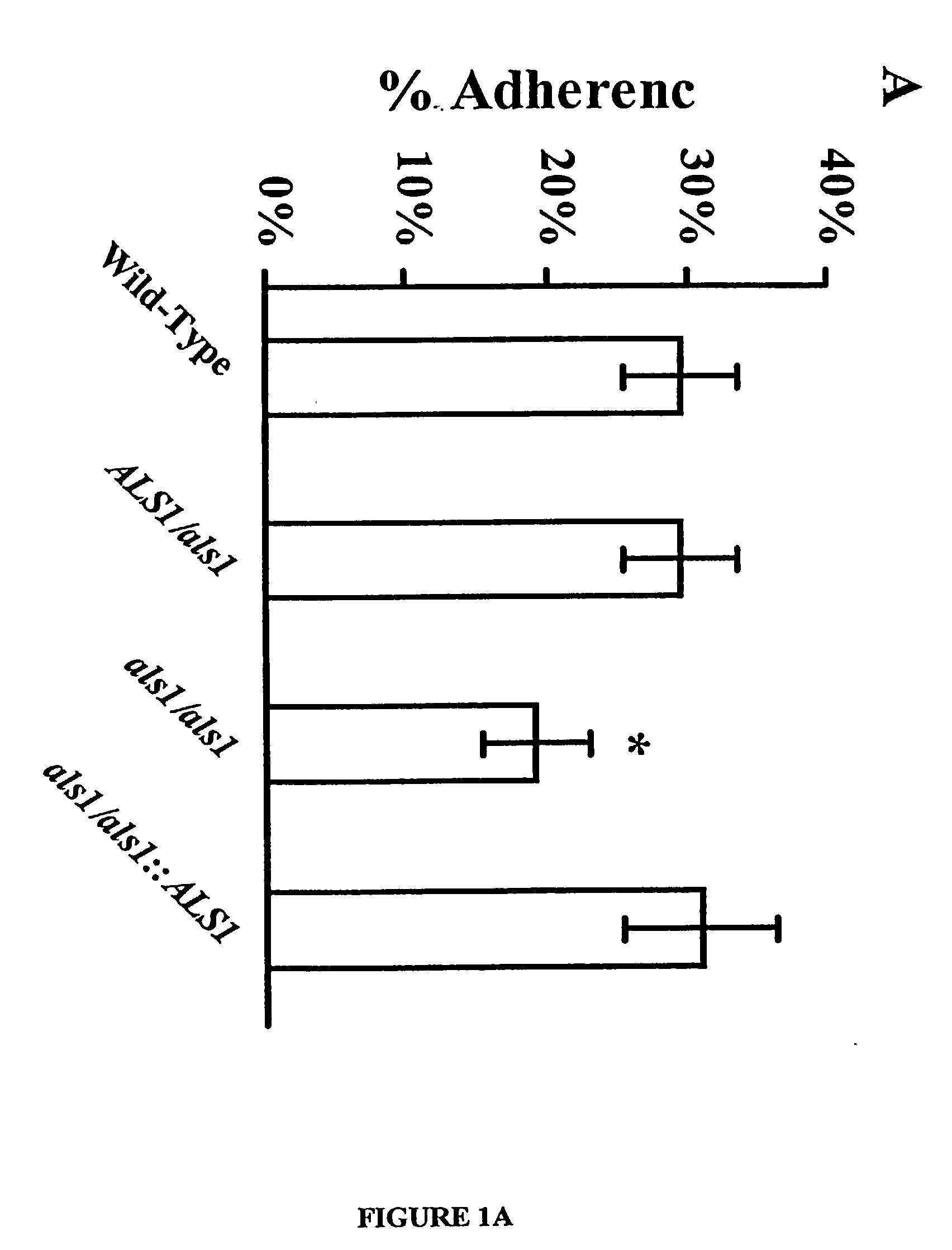
A Candida albicans bloodstream infections cause significant morbidity and mortality in hospitalized patients. Filament formation and adherence to host cells are critical virulence factors of C. albicans. Multiple filamentation regulatory pathways have been discovered, however the downstream effectors of these regulatory pathways remain unknown. The cell surface proteins in the ALS group are downstream effectors of the filamentation regulatory pathway. Particularly, Als1p mediates adherence to endothelial cells in vitro and is required for virulence. The blocking of adherence by the organism is described resulting from the use of a composition and method disclosed herein. Specifically, a pharmaceutical composition comprised of a gene, gene product, or specific antibody to the ALS gene family is administered as a vaccine to generate an immune response capable of blocking adherence of the organism.
Owner:LOS ANGELES BIOMEDICAL RES INST AT UCLA HARBOR MEDICAL CENT +1
Species-specific, genus-specific and universal DNA probes and amplification primers to rapidly detect and identify common bacterial and fungal pathogens and associated antibiotic resistance genes from clinical specimens for diagnosis in microbiology laboratories
InactiveUS20040185478A1Reduce usageDetermine rapidly the bacterial resistance to antibioticsMicrobiological testing/measurementFermentationBacteroidesNeisseria meningitidis
DNA-based methods employing amplification primers or probes for detecting, identifying, and quantifying in a test sample DNA from (i) any bacterium, (ii) the species Streptococcus agalactiae, Staphylococcus saprophyticus, Enterococcus faecium, Neisseria meningitidis, Listeria monocytogenes and Candida albicans, and (iii) any species of the genera Streptococcus, Staphylococcus, Enterococcus, Neisseria and Candida are disclosed. DNA-based methods employing amplification primers or probes for detecting, identifying, and quantifying in a test sample antibiotic resistance genes selected from the group consisting of blatem, blarob, blashv, blaoxa, blaZ, aadB, aacC1, aacC2, aacC3, aacA4, aac6'-lla, ermA, ermB, ermC, mecA, vanA, vanB, vanC, satA, aac(6')-aph(2''), aad(6'), vat, vga, msrA, sul and int are also disclosed. The above microbial species, genera and resistance genes are all clinically relevant and commonly encountered in a variety of clinical specimens. These DNA-based assays are rapid, accurate and can be used in clinical microbiology laboratories for routine diagnosis. These novel diagnostic tools should be useful to improve the speed and accuracy of diagnosis of microbial infections, thereby allowing more effective treatments. Diagnostic kits for (i) the universal detection and quantification of bacteria, and / or (ii) the detection, identification and quantification of the above-mentioned bacterial and fungal species and / or genera, and / or (iii) the detection, identification and quantification of the above-mentioned antibiotic resistance genes are also claimed.
Owner:GENEOHM SCI CANADA
Composition comprising a Lactobacillus pentosus strain and uses thereof
A novel probiotic strain termed Lactobacillus pentosus NCIMB 41114 is capable of preventing growth of pathogenic microorganisms in the GI tract. In particular, this bacterium inhibits Candida overgrowth, and can be employed to prevent and treat associated diseases, including IBS and thrush.
Owner:READING SCHOOL OF FOOD BIOSCI FOOD MICROBIAL SCI UNIT UNIV OF
Method and formulation for treating candidiasis using morinda citrifolia
InactiveUS7014873B2Good curative effectPrevent CandidiasisBiocideAnimal repellantsFruit juiceMonilinia laxa
The present invention features a novel use of processed ingredients from the Indian mulberry plant, and particularly a novel use of one or more processed Morinda citrifolia-based naturaceutical formulations comprising one or more of a processed Morinda citrifolia fruit juice, puree juice, oil or oil extract, dietary fiber, alcohol extract, etc., for inhibiting and preventing the overgrowth of Candida fungus and for treating Candidiasis and its associated symptoms.
Owner:TAHITIAN NONI INT INC
High-efficiency sterilization laundry detergent
InactiveCN103805367AImprove decontamination abilityEasy to rinseNon-ionic surface-active compoundsSurface-active non-soap compounds and soap mixture detergentsSURFACTANT BLENDStaphylococcus aureus bacteria
The invention discloses a high-efficiency sterilization laundry detergent. The high-efficiency sterilization laundry detergent comprises the following components in parts by weight: 0.5-6 parts of traditional Chinese medicine bactericide, 6-31 parts of anionic surfactant, 12-43 parts of nonionic surfactant, 0.06-1 part of thickening agent and other additives and 40-60 parts of deionized water, wherein a preparation method of the traditional Chinese medicine bactericide comprises the following steps: mixing folium artemisiae argyi, artemisia apiacea, radix scutellariae, pomegranate bark and folia perillae acutae which are equal in weight, decocting, filtering, concentrating filtrate to 1-1.5g / ml, and carrying out autoclaved sterilization. The high-efficiency sterilization laundry detergent prepared from the traditional Chinese medicine bactericide has a good inhibition effect on staphylococcus aureus, escherichia coli and candida albicans and strong cleansing power, and is rich in foam and easy to rinse.
Owner:刘菊
Species-specific, genus-specific and universal DNA probes and amplification primers to rapidly detect and identify common bacterial and fungal pathogens and associated antibiotic resistance genes from clinical specimens for diagnosis in microbiology laboratories
InactiveUS20030049636A1Low costRapid positioningMicrobiological testing/measurementFermentationBacteroidesNeisseria meningitidis
DNA-based methods employing amplification primers or probes for detecting, identifying, and quantifying in a test sample DNA from (i) any bacterium, (ii) the species Streptococcus agalactiae, Staphylococcus saprophyticus, Enterococcus faecium, Neisseria meningitidis, Listeria monocytogenes and Candida albicans, and (iii) any species of the genera Streptococcus, Staphylococcus, Enterococcus, Neisseria and Candida are disclosed. DNA-based methods employing amplification primers or probes for detecting, identifying, and quantifying in a test sample antibiotic resistance genes selected from the group consisting of blatem, blarob, blashv, blaoxa, blaZ, aadB, aacC1, aacC2, aacC3, aacA4, aac6'-lla, ermA, ermB, ermC, mecA, vanA, vanB, vanC, satA, aac(6')-aph(2''), aad(6'), vat, vga, msrA, sul and int are also disclosed. The above microbial species, genera and resistance genes are all clinically relevant and commonly encountered in a variety of clinical specimens. These DNA-based assays are rapid, accurate and can be used in clinical microbiology laboratories for routine diagnosis. These novel diagnostic tools should be useful to improve the speed and accuracy of diagnosis of microbial infections, thereby allowing more effective treatments. Diagnostic kits for (i) the universal detection and quantification of bacteria, and / or (ii) the detection, identification and quantification of the above-mentioned bacterial and fungal species and / or genera, and / or (iii) the detection, identification and quantification of the above-mentioned antibiotic resistance genes are also claimed.
Owner:BERGERON MICHEL G +3
Methods and materials for the production of organic products in cells of Candida species
InactiveUS7141410B2Good conditionUse in synthesisSugar derivativesMicroorganismsHeterologousBiotechnology
The present invention relates to biocatalysts that are cells, optimally of the Crabtree-negative phenotype, comprising expression vectors encoding genes heterologous to the cell that enable increased production of organic products. More specifically, the invention relates to genetically modified Candida cells, methods for making the Candida cells, and their use in production of organic products, particularly lactic acid.
Owner:CARGILL INC
Method for preventing and/or treating infections, colonisations, or illnesses related to staphylococcus aureus, pseudomonas aeruginosa, streptococcus pyogenes, enterococcus faecium, enterobacter cloacae, proteus mirabilis, bacteroides fragilis, staphylococcus epidermidis, propionibacterium acnes, candida albicans and/or malassezia furfur
The subject matter of the present invention is a bacteria or mix of bacteria having an antagonistic activity with respect to stains of S. aureus, P. aeruginosa, Streptococcus pyogenes, Enterococcus faecium, Enterobacter cloacae, Proteus mirabilis, Bacteroides fragilis Staphylococcus epidermidis, Propionibacterium acnes, Candida albicans and / or Malassezia furfur as well as the use thereof in the treatment and / or prevention of infections or colonisations related to those pathogens. The invention pertains to care products containing one or more non-pathogenic antagonistic strains intended to prevent and / or treat infections or colonisations on skin, wounds, mucous membranes and appendages.
Owner:URGO RECH INNOVATION & DEVEMENT
Microorganism bacterium agent for straw and excrement mixed composting
ActiveCN103232944AWell mixedPromote the process of mixed aerobic compostingFungiBio-organic fraction processingFecesPseudomonas putida
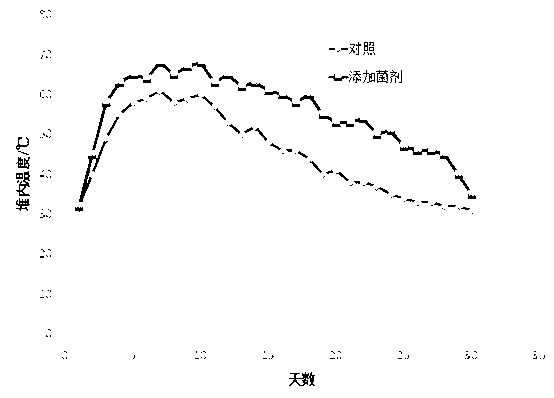
The invention discloses a microorganism bacterium agent for straw and excrement mixed composting. The microorganism bacterium agent is composed of the following strains: (2-6)*10<10>cfu of hair mould per gram, (2-6)*10<10>cfu of trichoderma virens per gram, (2-6)*10<10>cfu of geotrichum candidum per gram, (2-8)*10<9>cfu of saccharomyces cerevisiae per gram, (3-5)*10<11>cfu of bacillus thermophilus per gram, (2-6)*10<11>cfu of lactobacillus thermophilus per gram, (2-8)*10<10>cfu of white rot fungi per gram, (2-8)*10<10>cfu of monilinia fructicola per gram, (3-7)*10<8>cfu of pseudomonas putida per gram, (2-6)*10<11>cfu of nitrosococcus per gram, (1-4)*10<6>cfu of green algae per gram and (2-5)*10<7>cfu of blue-green algae per gram. The microorganism bacterium agent can promote a straw and excrement mixed aerobatic composting progress, shorten a composting period, improve a composting rotten degree, accelerate biodegradation of organic matters, alleviate ozone generation and diffusion during composting and reduce loss of nutrient substances such as nitrogen, can be applied to composting of different straw and excrement raw materials for composting enterprises and is used for producing bio-organic fertilizers.
Owner:山东土木启生物科技有限公司
Methods and compositions for detecting and identifying species of Candida
ActiveUS20080102449A1Avoid disadvantagesMicrobiological testing/measurementFermentationForward primerNucleotide
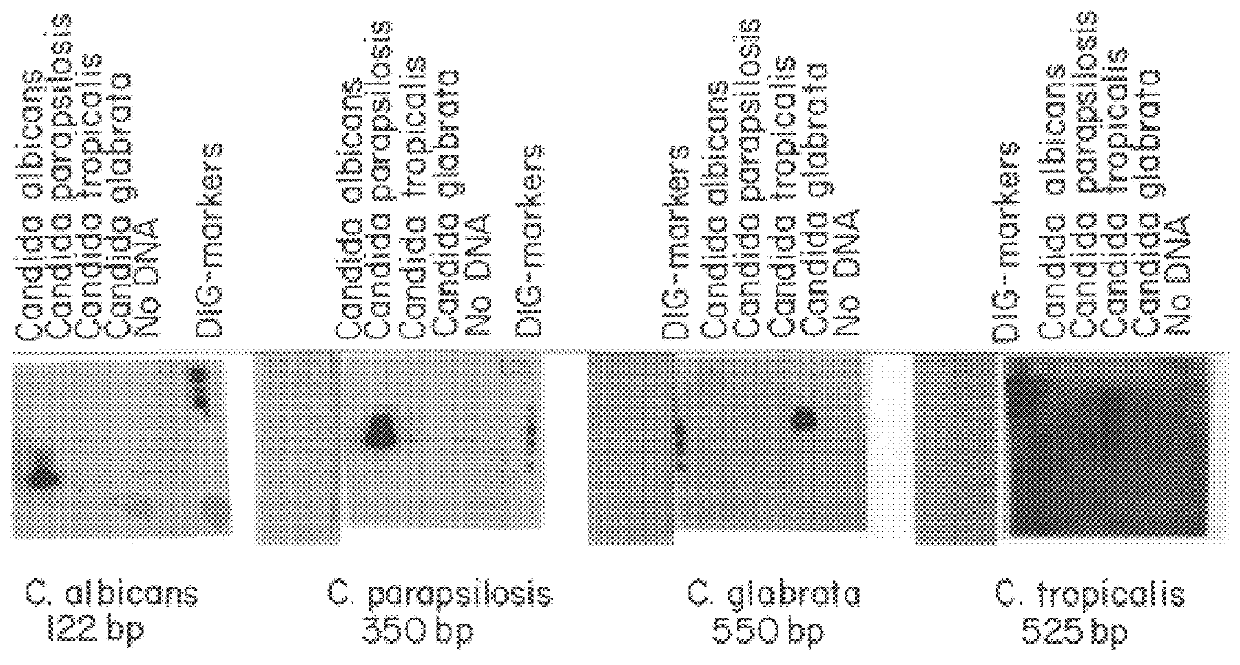
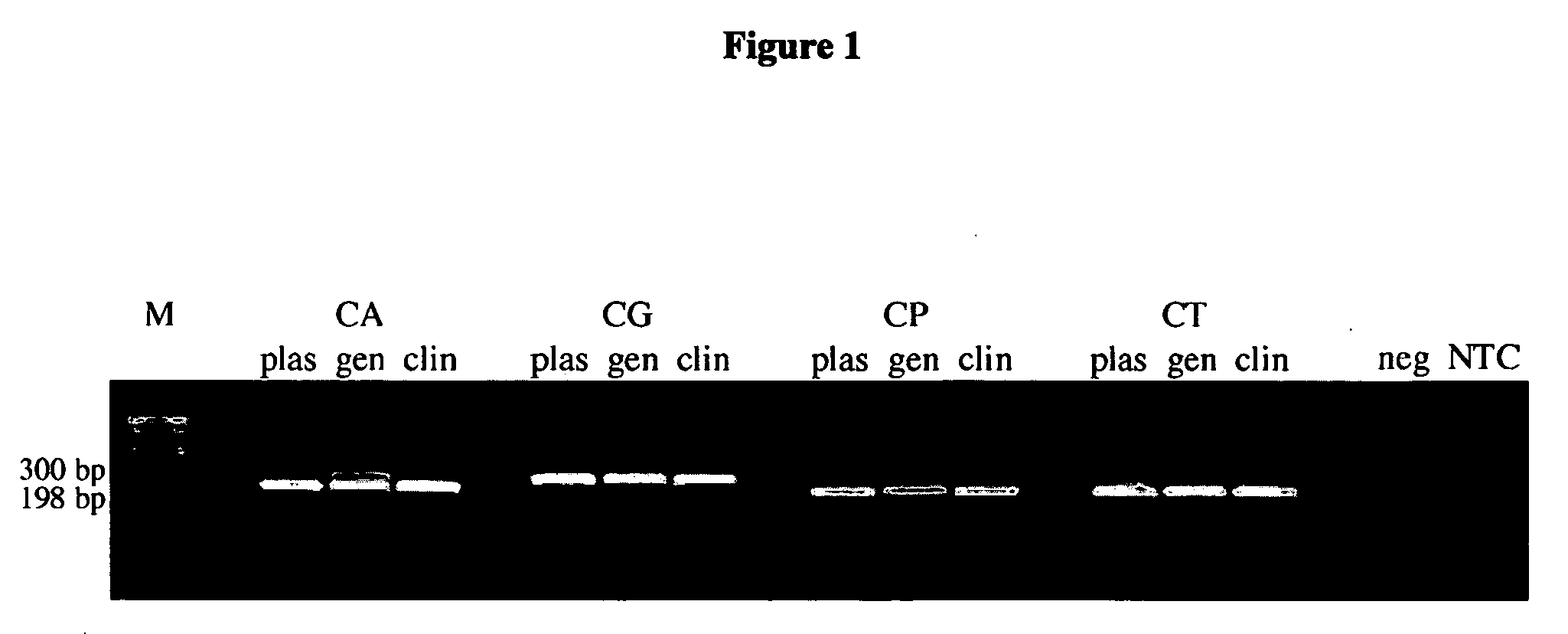
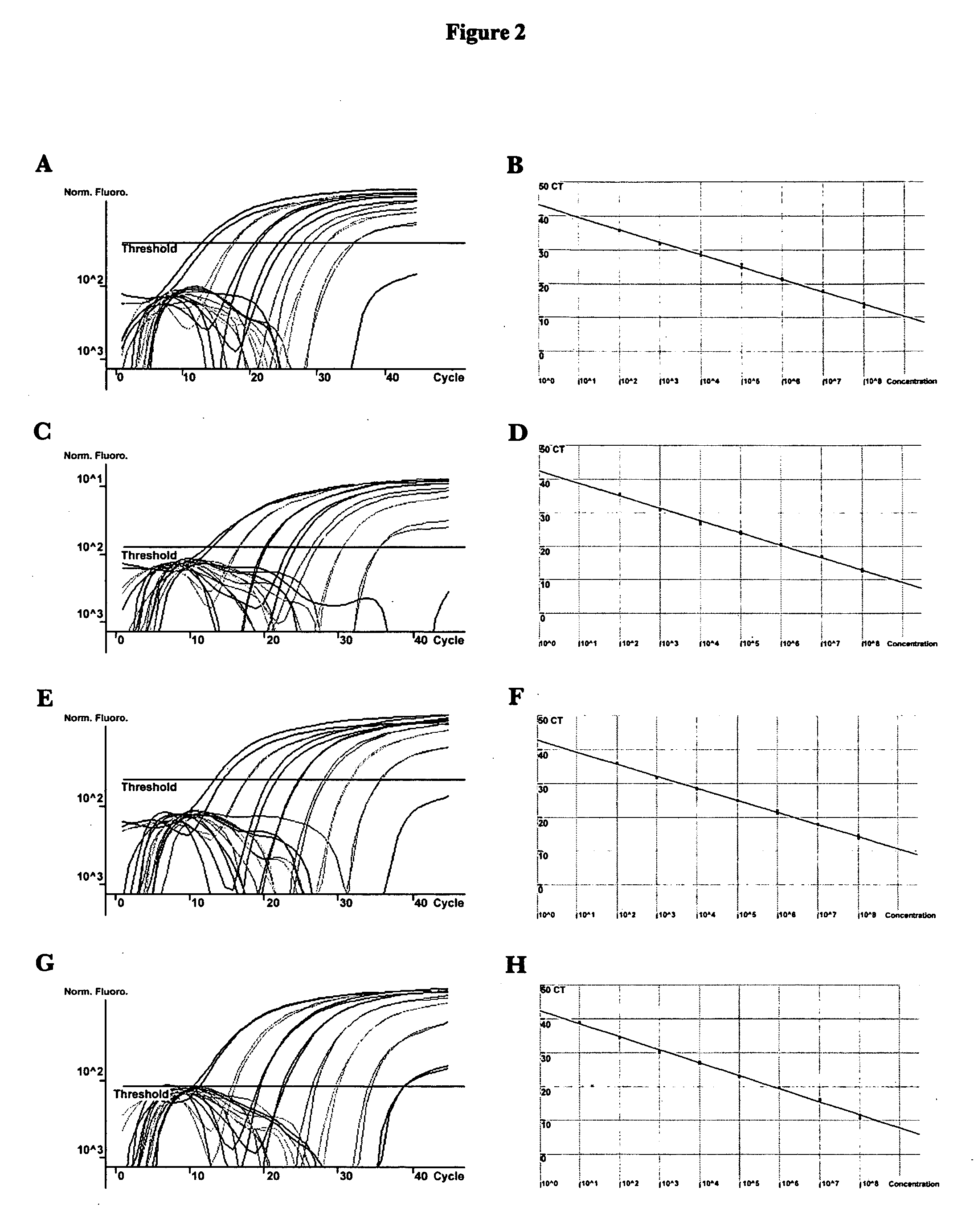
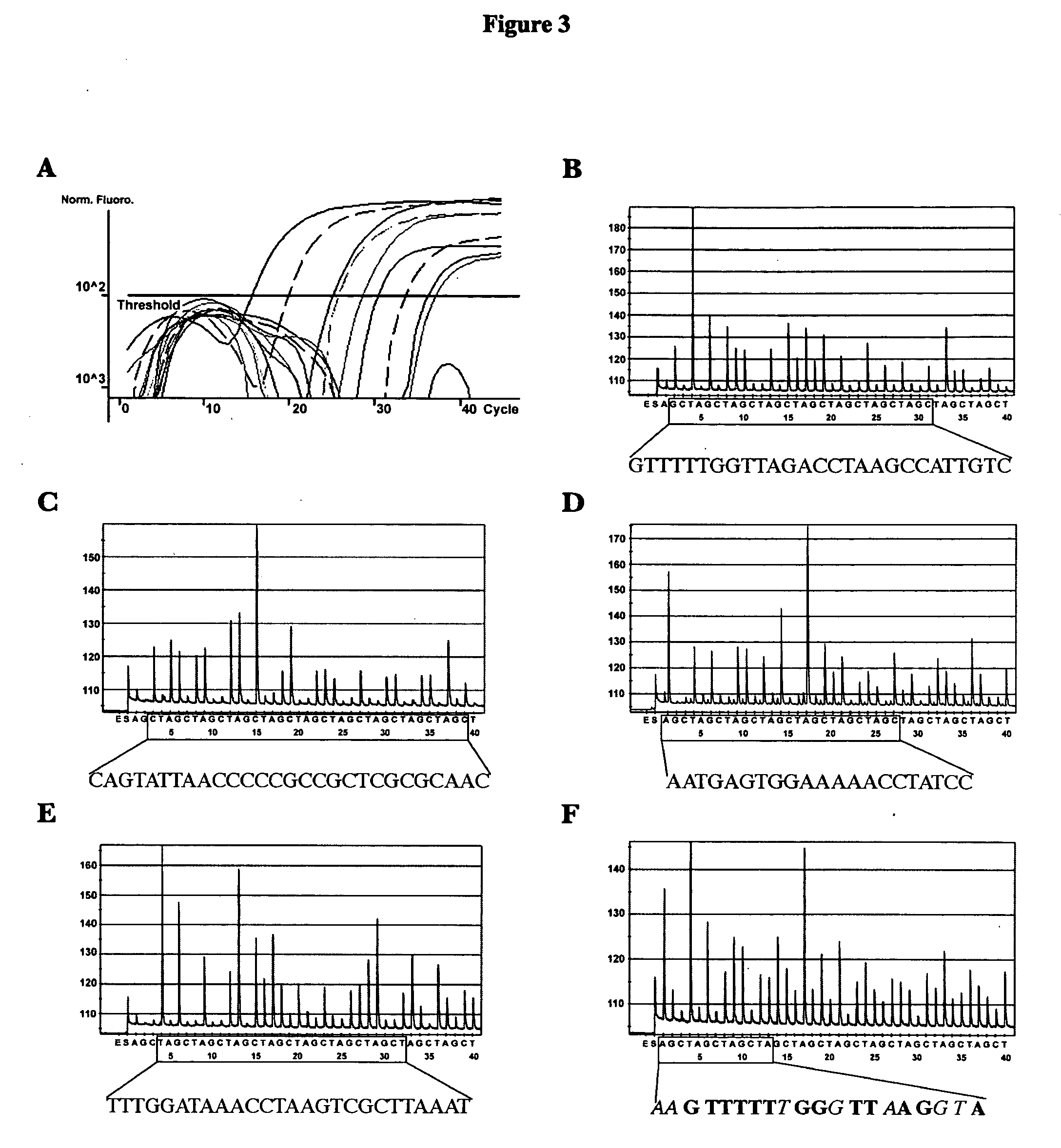
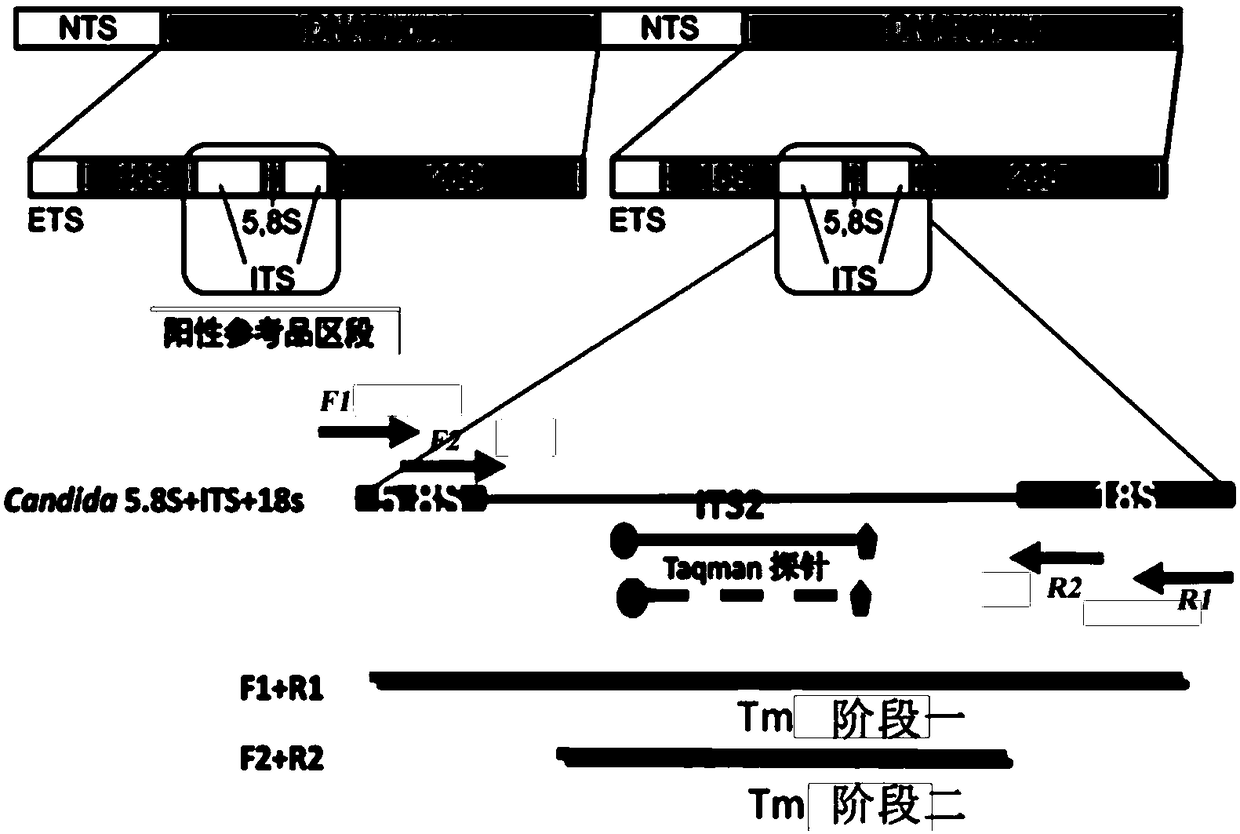
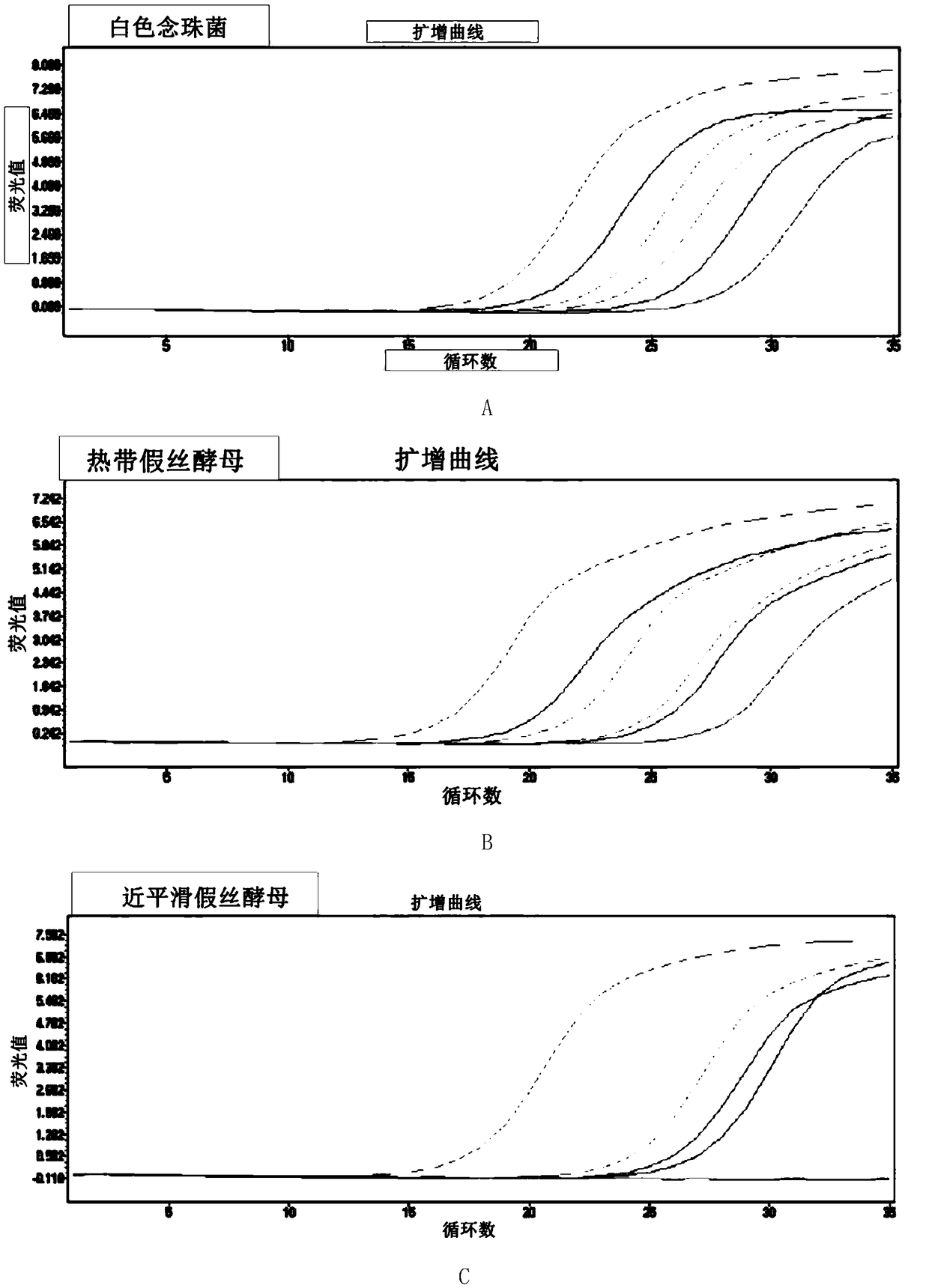
Methods and compositions useful in the detection and identification of species of Candida are disclosed. These species include Candida albicans, Candida glabrata, Candida parapsilosis, and Candida tropicalis, each of which is a causative agent for vaginal candidiasis. The compositions of the invention are combinations of oligonucleotides. These oligonucleotides include pairs of forward and reverse primers for polymerase chain reactions, wherein each primer pair is capable of priming the synthesis of an amplicon specific to one of Candida albicans, Candida glabrata, Candida parapsilosis, and Candida tropicalis, but preferably is not capable of priming the synthesis of an amplicon specific to any of the other three species. In preferred embodiments, the forward primers of the primer pairs have identical sequences, while each reverse primer of the primer pairs has a unique sequence relative to all of the other reverse primers; or the reverse primers of the primer pairs have identical sequences, while each forward primer of the primer pairs has a unique sequence relative to all of the other forward primers. These unique primer sequences account for the species specificity of the resultant amplicons. The oligonucleotides also include probes capable of detecting these amplicons, and sequencing primers for determining, in primer extension reactions, the nucleotide sequences contained within the amplicons. In preferred methods of the invention, a biological sample is tested for the presence of at least one isolate of Candida albicans, Candida glabrata, Candida parapsilosis, and Candida tropicalis by isolating nucleic acid from the sample, attempting a polymerase chain reaction in a mixture containing this nucleic acid and a plurality of these primer pairs, ascertaining whether any amplicon is produced in the mixture using an oligonucleotide probe, and determining the sequence of any resultant amplicon using the sequencing primers. The detection of an amplicon indicates that the sample contains at least one isolate of Candida albicans, Candida glabrata, Candida parapsilosis, or Candida tropicalis, and the nucleotide sequence data is used to determine which of these four Candida species is present.
Owner:MEDICAL DIAGNOSTIC LAB
Biochip for detecting pathogenesis fungus
InactiveCN1696311AReal-time detectionSensitive detectionMicrobiological testing/measurementMonilinia laxaCandida famata
A biochip for detecting the pathogenic funguses features that the DNA probes respectively for detecting one of 20 pathogenic funguses including Candida albicans, Aspergillus fumigatus, etc are immobilized on the glass plate, silicon chip, or high-molecular material.
Owner:INST OF HYGIENE & ENVIRONMENTAL MEDICINE PLA ACAD OF MILITARY MEDICAL
Polyhexamethylene guanidine propionate disinfectant and preparation method thereof
InactiveCN101574087AImprove the bactericidal effectEnhance long-term antibacterial effectBiocideDead animal preservationEscherichia coliNon toxicity
The invention provides a polyhexamethylene guanidine propionate disinfectant and a preparation method thereof. The polyhexamethylene guanidine propionate disinfectant is a hydrophilic solution prepared by the following steps: adding polyhexamethylene guanidine propionate, citric acid and surfactant to water and uniformly stirring the mixture at normal temperature. The polyhexamethylene guanidine propionate disinfectant has the advantages of broad spectrum of disinfection, quick results, long-time disinfection capability, high safety, non-toxicity, environmental protection, high stability, easy application, quick infiltration and volatilization, no coloration, easy cleaning, and the like. The polyhexamethylene guanidine propionate disinfectant has strong sterilization functions for colon bacillus, staphylococcus aureus, candida albicans, and the like with two-year effective periods. The polyhexamethylene guanidine propionate disinfectant can be used for disinfection of skins, wound surfaces, hands, air, departments of gynecology and urology, drinking water, and the like, thereby being a novel and ideal disinfectant.
Owner:铜陵高聚生物科技有限公司
Traditional Chinese medicine antibiotic skincare hand cleanser and preparation method thereof
InactiveCN102423291AGood antibacterial effectDelicate and not dryCosmetic preparationsToilet preparationsMonilinia laxaIrritation
The invention relates to a traditional Chinese medicine antibiotic skincare hand cleanser prepared from mixed materials of a traditional Chinese medicine extract, a surfactant, an antiseptic, glycerin, an essence and water. The traditional Chinese medicine extract is prepared from raw materials of, by weight: 35-45 parts of lightyellow sophora root, 15-25 parts of amur corktree bark, 15-25 parts of Chinese gentian, 5-15 parts of common cnidium fruit, and 5-15 parts of belvedere fruit. The raw materials are extracted by using water, and the density d of the obtained traditional Chinese medicine extract is 1.5-1.8g / ml. The traditional Chinese medicine antibiotic skincare hand cleanser provided by the invention has certain inhibitive effects against escherichia coli, staphylococcus aureus, and candida albicans, and provides certain preventive effects against eczema, skin pruritus, and the like. The hand cleanser is also advantaged in small dosage, high decontamination capability, good sterilization effect, and low irritation. The hands are delicate and not dry after washing. According to the invention, the formula of the hand cleanser is simple. With the hand cleanser, modern market requirements on naturalness, environment-friendliness, and high antibiotic efficiency can be satisfied.
Owner:SHANXI ZHENDONG PHARMA
Plant lactobacillus strain and its application
InactiveCN1888051AImprove cleanlinessImprove adhesionBacteriaBacteria material medical ingredientsEscherichia coliStaphylococcus cohnii
The present invention discloses one plant lactobacillus strain and its application. The plant lactobacillus, Lactobacillus plantarum L323 CGMCC No. 1329, is separated from Chinese pregnant woman's vaginal secretion, and has relatively high bacteriotasis on common vaginal pathogens, such as staphylococcus aureus, candida albicans, colibacillus and vaginal Gardnar bacillus, and acid producing and H2O2 producing capacity higher than other lactobacillus. The plant lactobacillus, Lactobacillus plantarum L323 CGMCC No. 1329, may be used in preparing vaginal microbial preparation for preventing and / or treating vaginal infectious diseases.
Owner:TIANYOUDA BIO ENG SCI & TECH BEIJING +1
Modified coconut oils with broad antimicrobial spectrum
InactiveUS20100016430A1Improve antibacterial propertiesAntibacterial agentsBiocideEscherichia coliCandida famata
Owner:MALAYSIAN AGRI RES & DEV INST MARDI
Antimicrobial hexapeptides
The invention encompasses hexapeptides consisting of alternating hydrophobic residues (B) at positions 2, 4, and 6, hydrophilic, charged residues (X) at positions 1 and 3, and a naphthylalanine (Nal), an aliphatic or aromatic residue (O) at position five, represented generally by the formula XBXBOB, which exhibit antimicrobial activity against infections caused by a variety of pathogens. These pathogens may include gram positive or negative bacteria, acid-fast bacteria such as mycobacteria, parasites, dermatophytes, or fungal pathogens. Typical fungal pathogens include Candida albicans and typical dermatophytes include Trichophyton rubrum and Trichophyton mentagrophytes. The hexapeptides of the present invention exhibit antifungal activity, antibacterial activity, desirable stability, and lack toxicity to the mammal receiving treatment.
Owner:HELIX BIOMEDIX INC
Oral antibacterial composition containing nisin and polylysine and preparation method thereof
InactiveCN105326656AImprove the bactericidal effectNo adverse reactionAntibacterial agentsCosmetic preparationsSide effectPorphyromonas gingivalis
The invention provides an oral antibacterial composition containing nisin and polylysine. According to the composition, the growth of oral pathogenic bacteria such as porphyromonas gingivalis, actinobacillus actinomycetemcomitans, streptococcus mutans, lactic acid bacillus, staphylococcus aureus and candida albicans can be effectively inhibited, and tooth decay, gingivitis, periodontitis, ozostomia and the like can be prevented; the composition can be prepared into toothpaste, mouth wash, refreshing and tasting chips and chewing gum which are used for preventing and treating oral diseases. Meanwhile, the invention provides a preparation method of the compositions such as the toothpaste, the mouth wash, the refreshing and tasting chips and the chewing gum. The oral antibacterial composition prepared through the method can prevent and treat various oral diseases and has no toxic and side effect and allergy or stimulation phenomenon on a human body.
Owner:SHANDONG ACADEMY OF PHARMACEUTICAL SCIENCES
Fluorescence quantitative PCR primer, probe and kit for detecting ordinary pathogenic fungi
InactiveCN104450936AOvercome limitationsSolve the problem of fungal infectionMicrobiological testing/measurementDNA/RNA fragmentationFluorescenceCandida tropicalis
The invention discloses a fluorescence quantitative PCR primer, a probe and a kit for detecting ordinary pathogenic fungi. A specific primer and a TaqMan probe are independently designed, and a fluorescence PCR detection method which can be used for simultaneously detecting 15 clinically ordinary pathogenic fungi, including 8 candida mycoderma bacteria (candida albicans, candida glabrata, candida parapsilosis, candida kefyr, candida sake, candida kruse, candida guilliermind and candida tropicalis), four aspergilli (aspergillus niger, aspergillus flavus, aspergillus terreus and aspergillus fumigates), cryptococcus, rhizopus oryzae and mucor circinelloides, is established. The method is relatively high in sensitivity and specificity, and has significant meanings for early diagnosis and treatment on invasive infections with fungi.
Owner:天津宝瑞生物技术有限公司
Herbal antiseptic containing chitosan
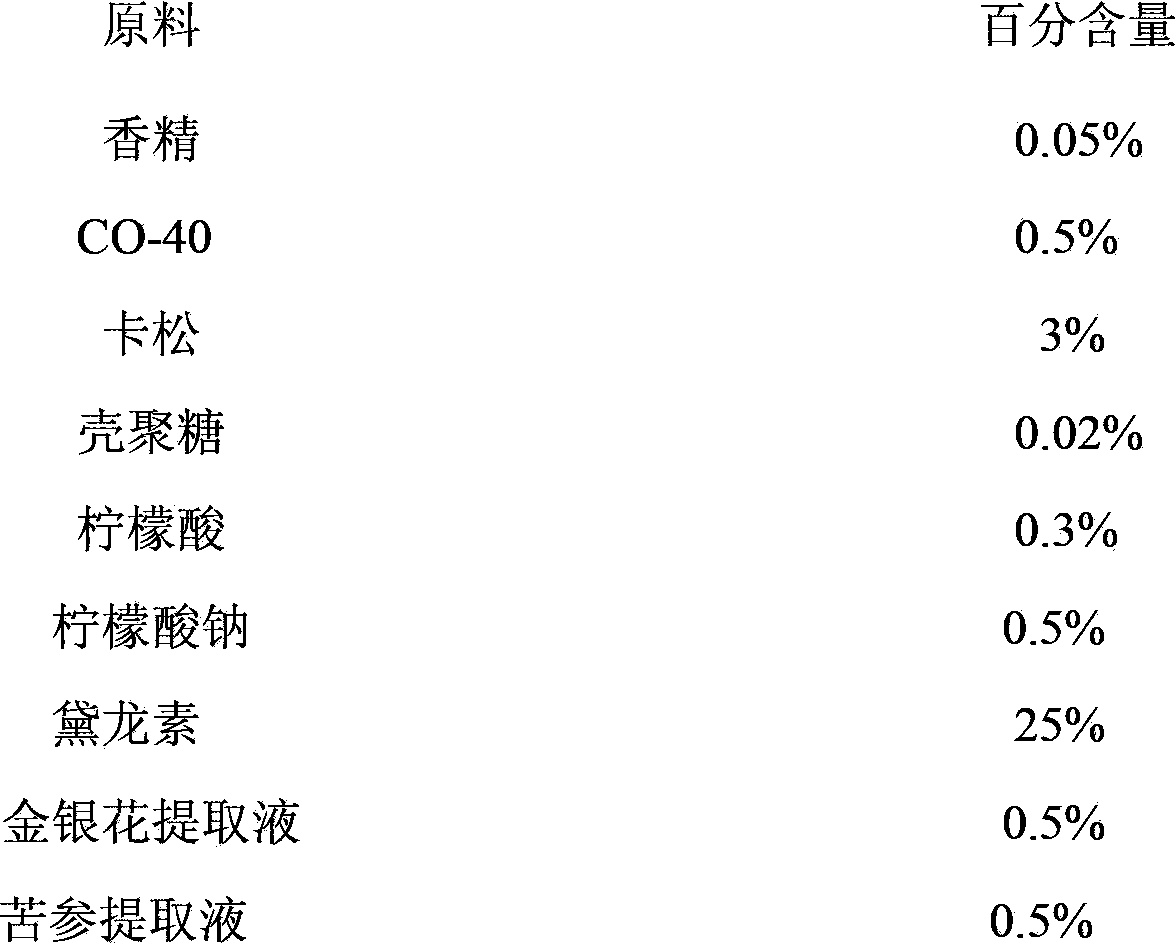
InactiveCN103766415AEnhanced inhibitory effectStable in natureBiocideFungicidesGlycerolAntiseptic solutions
The invention relates to a herbal antiseptic containing chitosan. The antiseptic comprises the following components in percentage by weight: 0.05% of essence, 0.5% of CO-400, 3% of kathon, 0.02% of chitosan, 0.3% of critic acid, 0.5% of sodium citrate, 25% of deloncide, 0.5% of honeysuckle flower extract, 0.5% of sophora flavescens extract, 1.5% of glycerin, and the balance being purified water, and the antiseptic is prepared by evenly mixing the components mentioned above. The herbal antiseptic has the advantages that: (1) the antiseptic solution has a weak acidic property, and the pH value of the solution is in a range of 4 to 7; (2) the antiseptic has a prominent and long-lasting inhibiting effect on bacteria and fungi, the candida albicans inhibiting rate is more than or equal to 90%, the staphylococcus aureus inhibiting rate is more than or equal to 90%, and the colibacillus inhibiting rate is more than or equal to 90%; (3) the antiseptic has a stable property, and does not disintegrate or change color.
Owner:TIANJIN SINOSH NEW MATERIAL TECH
Viscose fiber containing radix isatidis extract and preparation method of viscose fiber
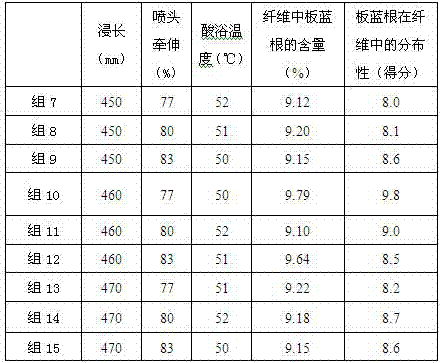
ActiveCN106929931AImprove uniformityShort stayArtificial filaments from viscoseMicroballoon preparationCandida albicansViscose
The invention provides viscose fiber containing a radix isatidis extract. According to the viscose fiber, the content of radix isatidis microcapsules is 9.1%-9.79%, the dry fracture strength of the fiber is 2.70cN / dtex-2.75cN / dtex, the wet fracture strength of the fiber is 1.70cN / dtex-1.75cN / dtex, the elongation at break of the fiber is 22%-23%, the antibacterial rate of the fiber to staphylococcus aureus is 99.0%-99.8%, the antibacterial rate of the fiber to Candida albicans is 96.6-98.8%, the antibacterial rate of the fiber to typhoid bacillus is 97.2%-99.1%, and the fiber has very strong effects of inhibiting and killing influenza A virus and influenza B virus. The invention further provides a preparation method of the viscose fiber containing the radix isatidis extract. The preparation method comprises a step of modifying the radix isatidis microcapsules. According to the preparation method, the loss of the radix isatidis microcapsules in the preparation process of the fiber is reduced, the dismantling backwashing rate of a coagulating bath filter is reduced, and the distribution uniformity of the radix isatidis microcapsules in the fiber is increased.
Owner:青岛邦特生态纺织科技有限公司
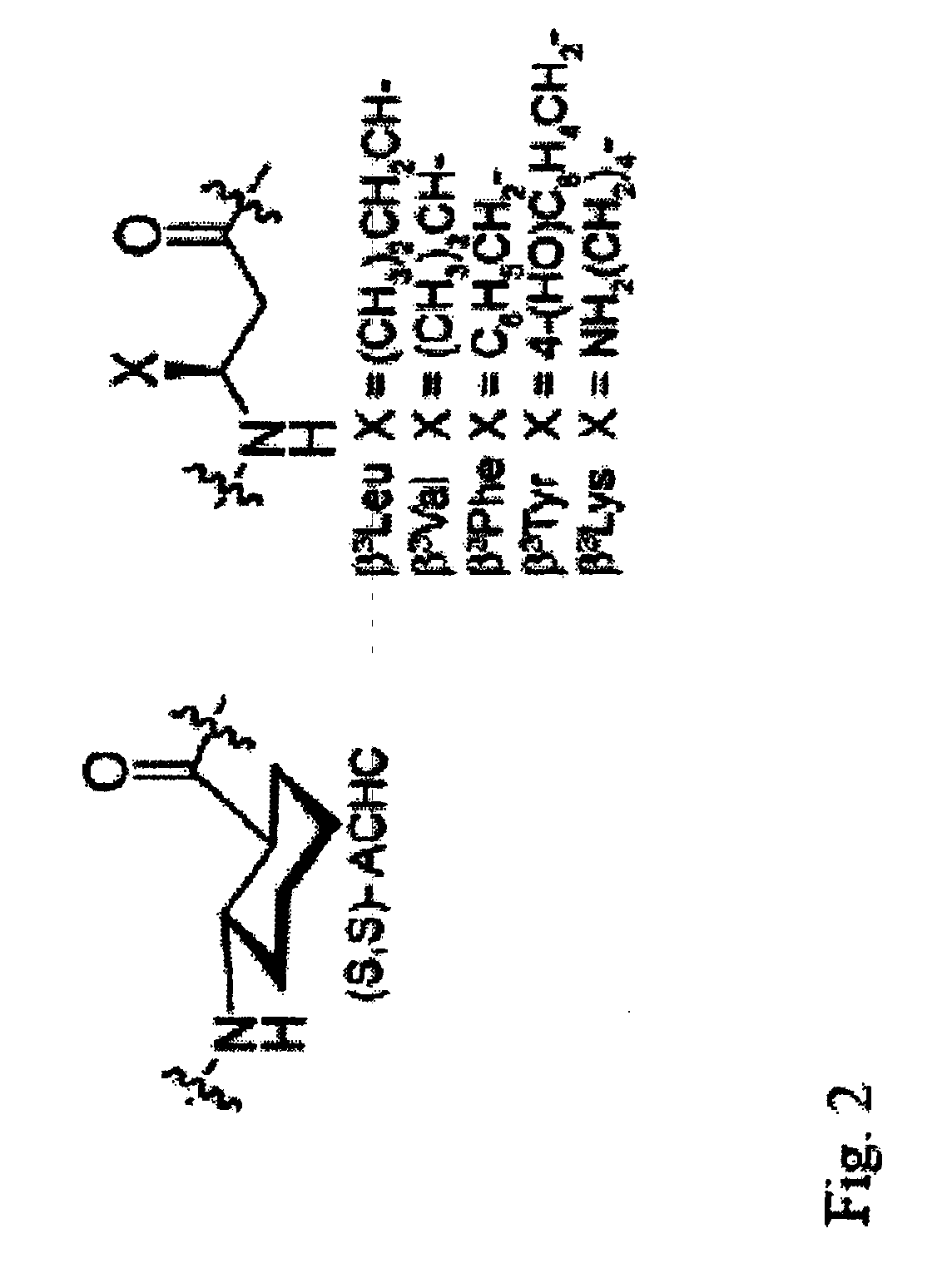
Beta-peptides with antifungal activity
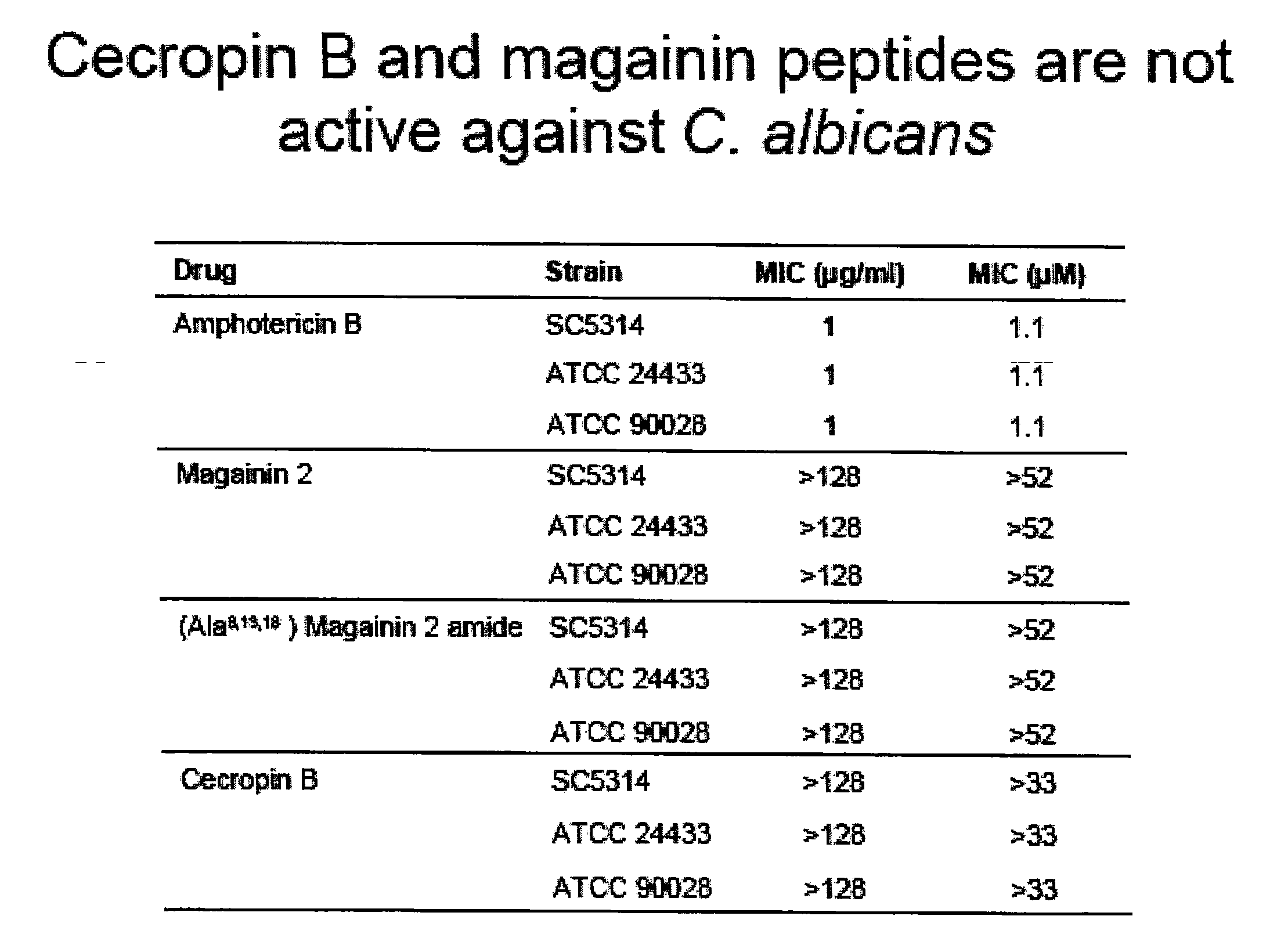
The present invention is directed to the design, synthesis and use of various β-peptides exhibiting antifungal activity. The β-peptides are relatively short in length, adopt globally amphiphilic conformations, and cause little lysis of human red blood cells at concentrations that kill Candida albicans, a common human fungal pathogen.
Owner:WISCONSIN ALUMNI RES FOUND
Preparation methods of sustained-release microcapsule and sustained-release composite membrane for inhibiting monilinia fructicola
The invention discloses preparation methods of a sustained-release microcapsule and a sustained-release composite membrane for inhibiting the monilinia fructicola. The preparation method of the sustained-release microcapsule comprises the following steps: the chitosan and the berberine are weighed, the mass ratio between the chitosan and the berberine is 9-11:0.22-0.28, the acetate solution is added, then the chitosan and the berberine are sufficiently swelled to obtain berberine-chitosan solution after stirring for dissolving, standing and air bubble removal, and the berberine-chitosan solution is added in the soybean oil containing span 80, stirred at the constant temperature of 40-50 DEG C and then emulsified for 28-32 min to form a W / O system; the glutaraldehyde solution is added for stirring for 2.5-3.5 hours, and the centrifuging is carried out so that the microcapsule is fully settled; and the preparation liquid on the upper layer is obliquely taken, the obtained microcapsule is washed with petroleum ether for 3-5 times and then dried for 23-25 hours under the vacuum at the temperature of 48-52 DEG C. The invention also provides the preparation method for the sustained-release composite membrane for inhibiting the monilinia fructicola. Both the microcapsule and the composite membrane have the sustained-release effects and have no toxic side effect to the environment and the plants since the berberine is a native compound.
Owner:BEIJING UNIV OF CHEM TECH
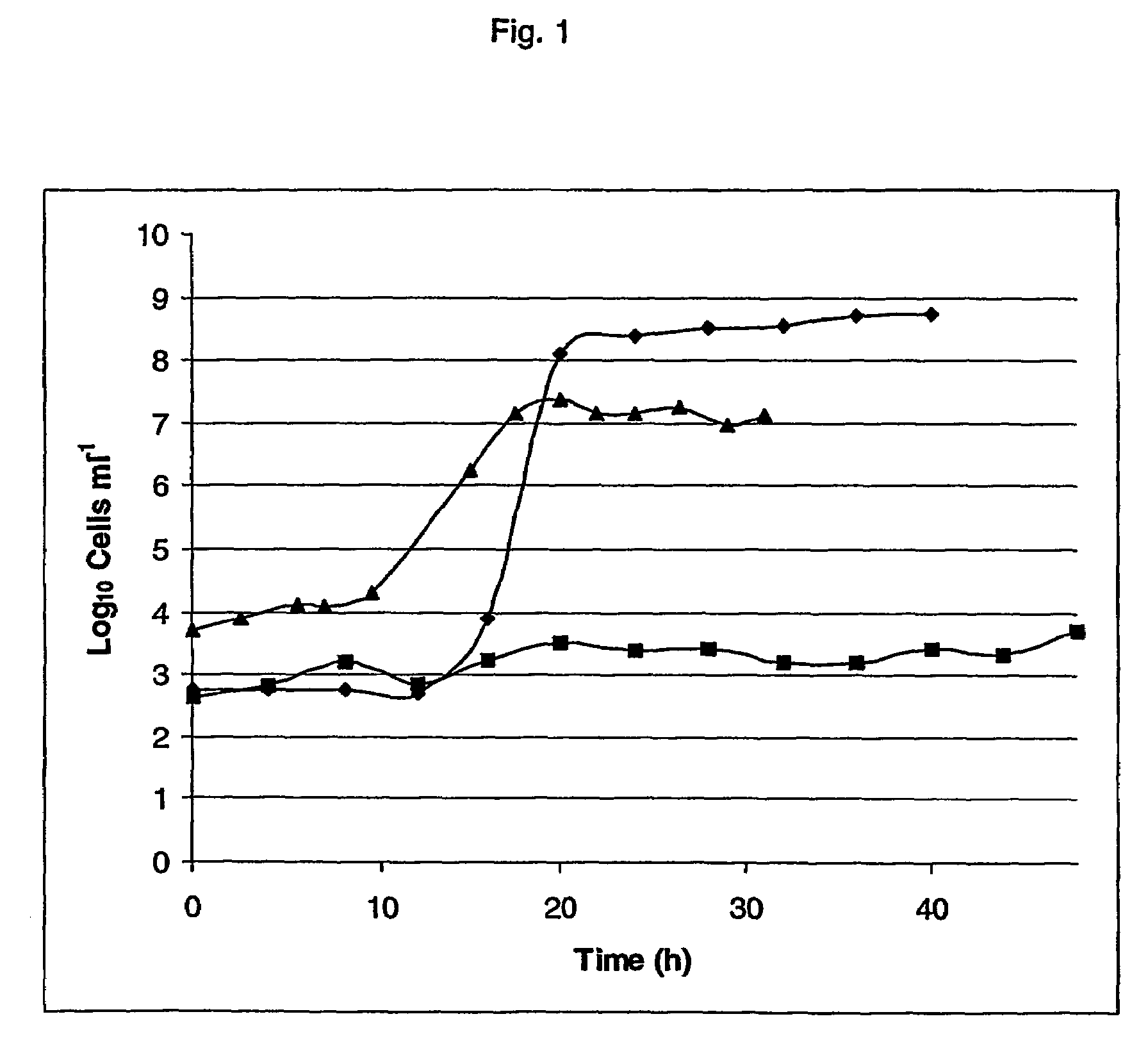
Processes for the bioconversion of a fermentable carbon source to 1,3-propanediol by a single microorganism
A process is provided for the bioconversion of a carbon substrate to 1,3-propanediol by a single organism utilizing microorganisms, such as, Citrobacter, Enterobacter, Clostridium, Klebsiella, Aerobacter, Lactobacillus, Aspergillus, Saccharomyces, Zygosaccharomyces, Pichia, Kluyveromyces, Candida, Hansenula, Debaryomyces, Mucor, Torulopsis, Methylobacter, Escherichia, Salmonella, Bacillus, Streptomyces and Pseudomonas, containing the genes encoding for an active glycerol or diol dehydratase enzyme by contacting these organisms with a carbon substrate under the appropriate fermentation conditions. Specifically, Citrobacter and, Klebsiella provide the source of exogenous genes for such active dehydratase enzyme.
Owner:EI DU PONT DE NEMOURS & CO +1
Metschnikowia pulcherrima inhibiting fungi and application thereof
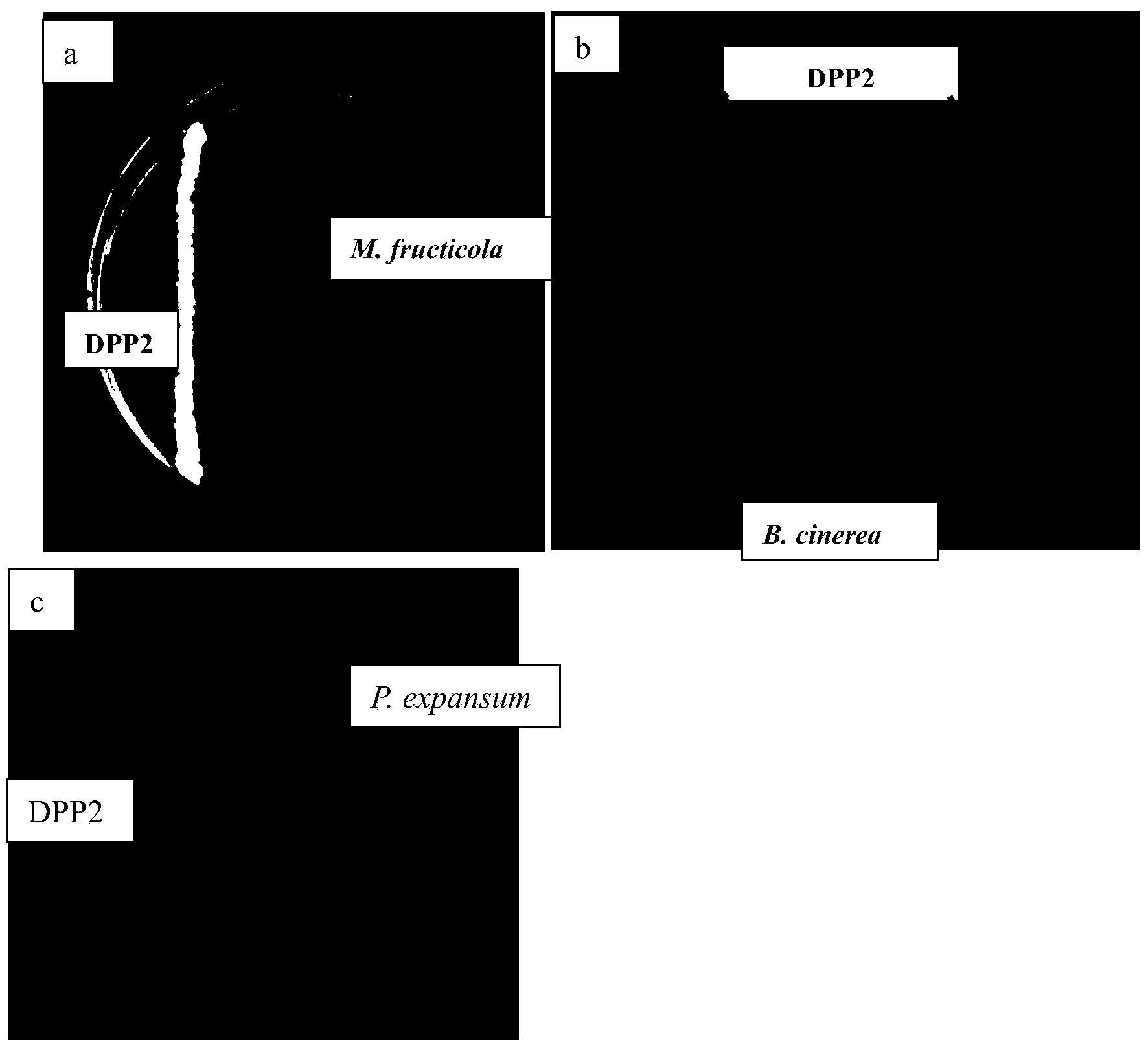
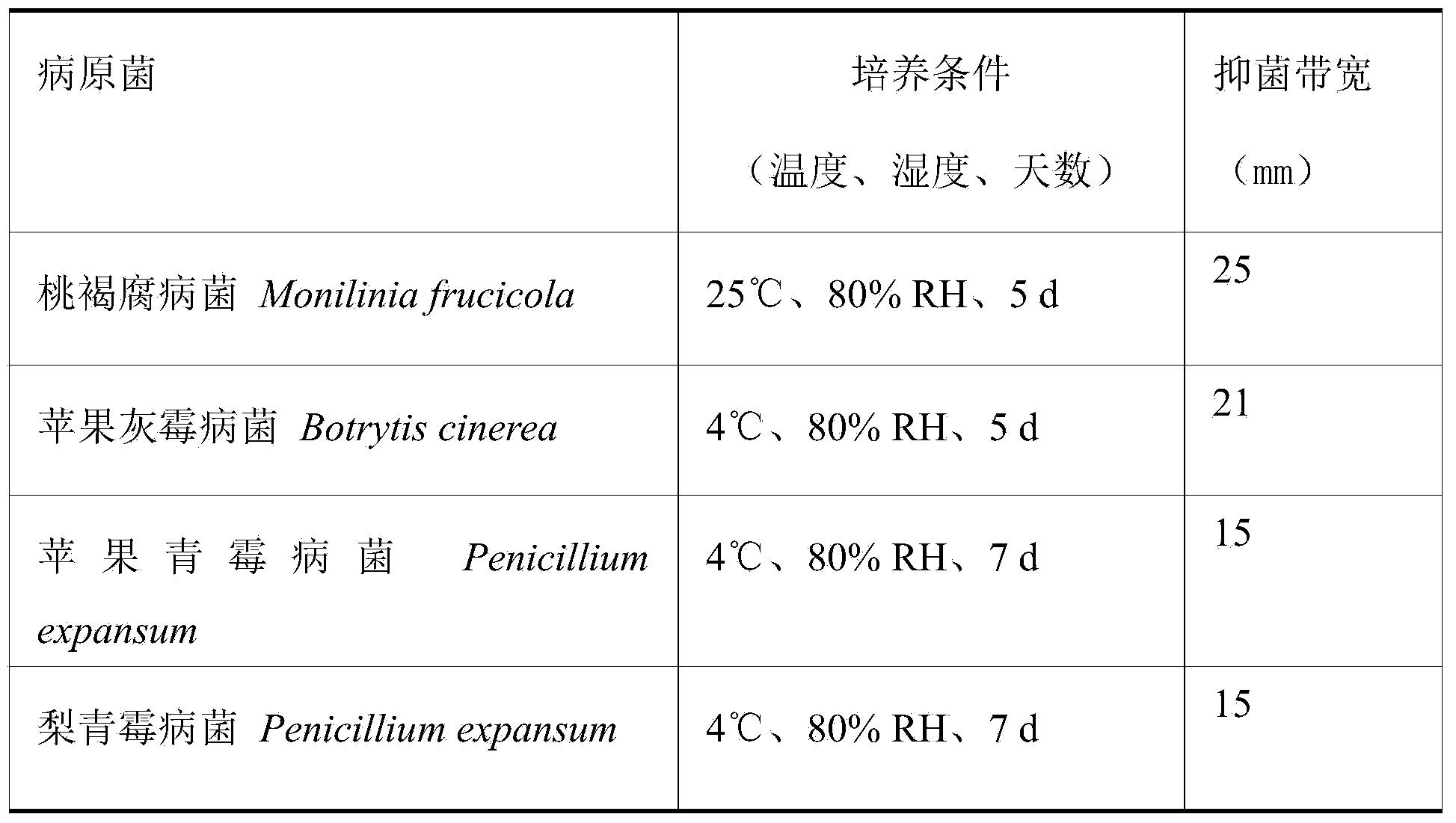
The invention discloses Metschnikowia pulcherrima DPP2 with the collection number of CGMCC No. 8178. The Metschnikowia pulcherrima DPP2 has an inhibiting effect on peach postharvest monilinia fructicola, apple postharvest botrytis cinerea, apple postharvest penicillium notatum and pear postharvest penicillium notatum, as well as the inhibiting effect on peach postharvest brown rot, apple postharvest gray mold, pear postharvest blue mold and apple postharvest blue mold. In the selected plant pathogenic fungi, the inhibiting effect on the peach monilinia fructicola and the apple botrytis cinerea is most obvious, the diameters of inhibition zones are more than 20mm respectively, and the Metschnikowia pulcherrima DPP2 has the advantages of stability, high efficiency and broad-spectrum antibacterial property, and can be applied to control of pathogenic bacteria or diseases.
Owner:BEIJING ACADEMY OF AGRICULTURE & FORESTRY SCIENCES
Biological antibacterial composition and application of biological antibacterial composition in cosmetics
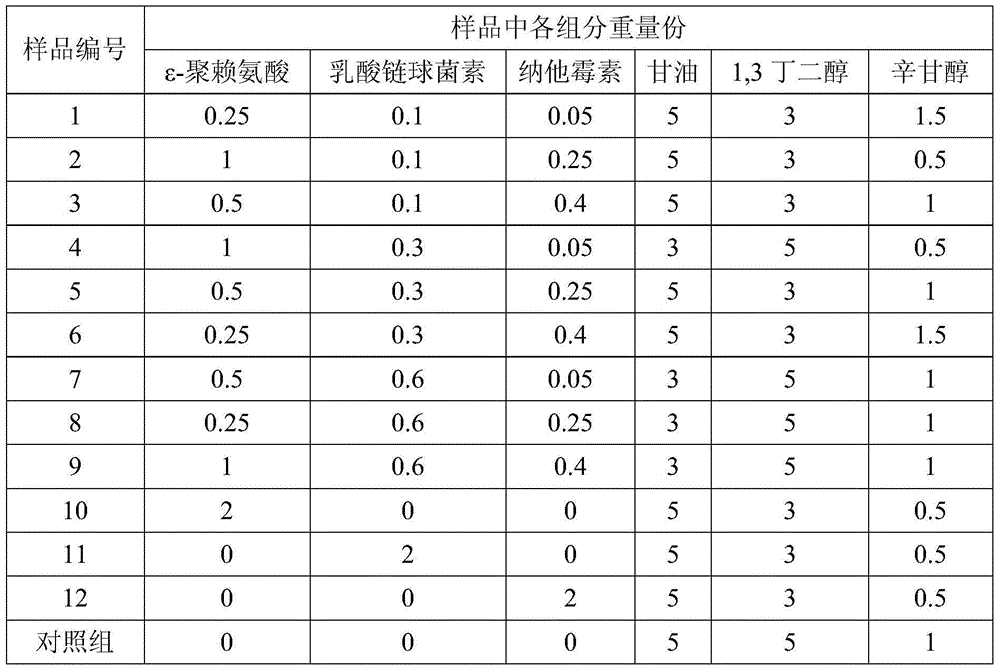
ActiveCN104958248AAdd lessGood antibacterial effectCosmetic preparationsToilet preparationsCaprylyl GlycolGlycerol
The invention provides a biological antibacterial composition and application of the biological antibacterial composition in cosmetics. The biological antibacterial composition comprises the following components in parts by weight: 0.25 to 1.0 part of epsilon-polylysine, 0.1 to 0.6 parts of nisin, 0.05 to 0.4 parts of natamycin, 3 to 8 parts of glycerin, 2 to 8 parts of 1,3-butanediol and 0.2 to 2 parts of caprylyl glycol. The composition provided by the invention has good antibacterial effect, particularly has a significant synergistic effect on inhibiting candida albicans, pseudomonas aeruginosa and aspergillus niger, can be used in skin care and washing cosmetics, and the additive amount of the original preservatives is reduced.
Owner:TIANJIN YU MEI JING GRP
Antibacterial soap and preparation method thereof
InactiveCN102329703AEfficient removalEfficient killingAlkali/ammonium soap compositionsEscherichia coliStaphylococcus aureus
The invention discloses antibacterial soap which is characterized by comprising the following raw material components: 5-15% of coconut oil, 10-20% of palm oil, 3-10% of olive oil, 15-30% of beef tallow, 5-15% of sodium hydroxide, 2-8% of liquorice water extract, 1-3% of sodium dodecyl benzene sulfonate and 3-8% of titanium dioxide. The antibacterial soap obtained in the invention can be directlyapplied to the skin to effectively eliminate and kill bacteria on the skin and can resist staphylococcus aureus, Pseudomonas aeruginosa, escherichia coli and Candida albicans simultaneously.
Owner:NINGXIA MEDICAL UNIV
Detection method of invasive fungus infection, detection kit and application
ActiveCN109055502ASimplify detectionSimplify operabilityMicrobiological testing/measurementDNA/RNA fragmentationCell freeFluorescence
The invention provides a fast multiplex PCR identification diagnosis detection method of invasive fungus infection based on cfDNA (cell free DNA). The method can be used for identifying the invasive infection caused by clinic common and high-incidence Candida albicans, Candida tropicalis, Candida parapsilosis, Candida krusei, Candida glabrata and Aspergillus fumigtus. An amplification primer is designed according to characteristic genome segments of each fungus category and species; a detection fluorescence probe of a strain can be distinguished according to the amplification segment design; the real time PCR can be performed on a sample to be tested; high sensitivity of nested PCR and high specificity and multi-target performance advantages of multiple fluorescent hybrid probe PCR are integrated for identifying the fungus strain. The invention also provides a PCR diagnosis kit for the invasive fungus infection and application thereof.
Owner:DIASYS DIAGNOSTIC SYST SHANGHAI
Biooxidation capabilities of Candida sp
Owner:COGNIS IP MANAGEMENT GMBH
Weissella cibaria XHR1 and applications thereof and weissella cibaria XHR1 containing pickled vegetable
ActiveCN105255787AImprove micro-ecological levelExpansion of industrial productionBacteriaMicroorganism based processesWeissella cibariaCandida famata
The invention provides a weissella cibaria XHR1 and applications thereof and a weissella cibaria XHR1 containing pickled vegetable. The preparation of the pickled vegetable is implemented through the steps of adding a culture solution of weissella cibaria XHR1 and bacterial powder of lactobacillus plantarum CICC 20764 into a pickled vegetable fermentation starting solution for pickling vegetables, so that the final concentration of weissella cibaria XHR1 is 10<6>-10<7> cfu / mL eventually, and then the final concentration of lactobacillus plantarum CICC 2076 is 10<5>-10<6> CFU / mL; and then, adding vegetables into the pickled vegetable fermentation starting solution, and carrying out vegetable pickling according to a traditional vegetable fermentation process, so that the pickled vegetable disclosed by the invention is obtained finally. According to the invention, prebiotic lactobacillus for inhibiting the growth of Candida albicans is applied to the fermentation of Sichuan pickled vegetables, so that the consumer group is wider, and the micro ecological level of alimentary canals of wide consumers can be improved, thereby inhibiting Candida albicans diseases from the perspective of public health.
Owner:XIHUA UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com