Method and kit for identifying and detecting babesia unidentified species
A detection kit, Babesia technology, applied in microorganism-based methods, biochemical equipment and methods, and microbial determination/inspection, etc. problems, to ensure a high degree of specificity, a wide range of use, and low skill requirements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1: Detection of Babesia mosoni in goat or sheep blood samples
[0025] 1. Blood Genomic DNA Extraction
[0026] Blood DNA was extracted with QIAamp DNA Mini Kit (Qiagen, Germany), and the specific operation steps were carried out according to the instructions.
[0027] 2. Primer Design
[0028] According to the RON2 gene sequence of Babesia mosoni (Lintan strain, Tianzhu strain, Ningxian strain and Hebei strain) Babesia undetermined species (Xinjiang strain and Dunhuang strain) released by GenBank, after sequence comparison analysis, within the species Conservative, inter-species variation sequence design cross-amplification primers, the amplification length is 150bp.
[0029] Replacement primer BXJ-5a: 5'-AAAGTTGAAACTCCCACACC-3'
[0030] Replacement primer BXJ-4s: 5'-CGATGCTCTGCTCCAAGGTA-3'
[0031] Cross primer BXJ-2a1s: 5'-
[0032] TTGATATACCGGAGGCCGATCCGGTTATCAACATTTGTGGCGAC-3'
[0033] Probe BXJ-2a: 5'-FITC-TTGATATACCGGAGGCCGATCC-3'
[0034] Probe B...
Embodiment 2
[0048] Example 2: Specific detection of cross primer amplification detection
[0049] 1. Blood Genomic DNA Extraction
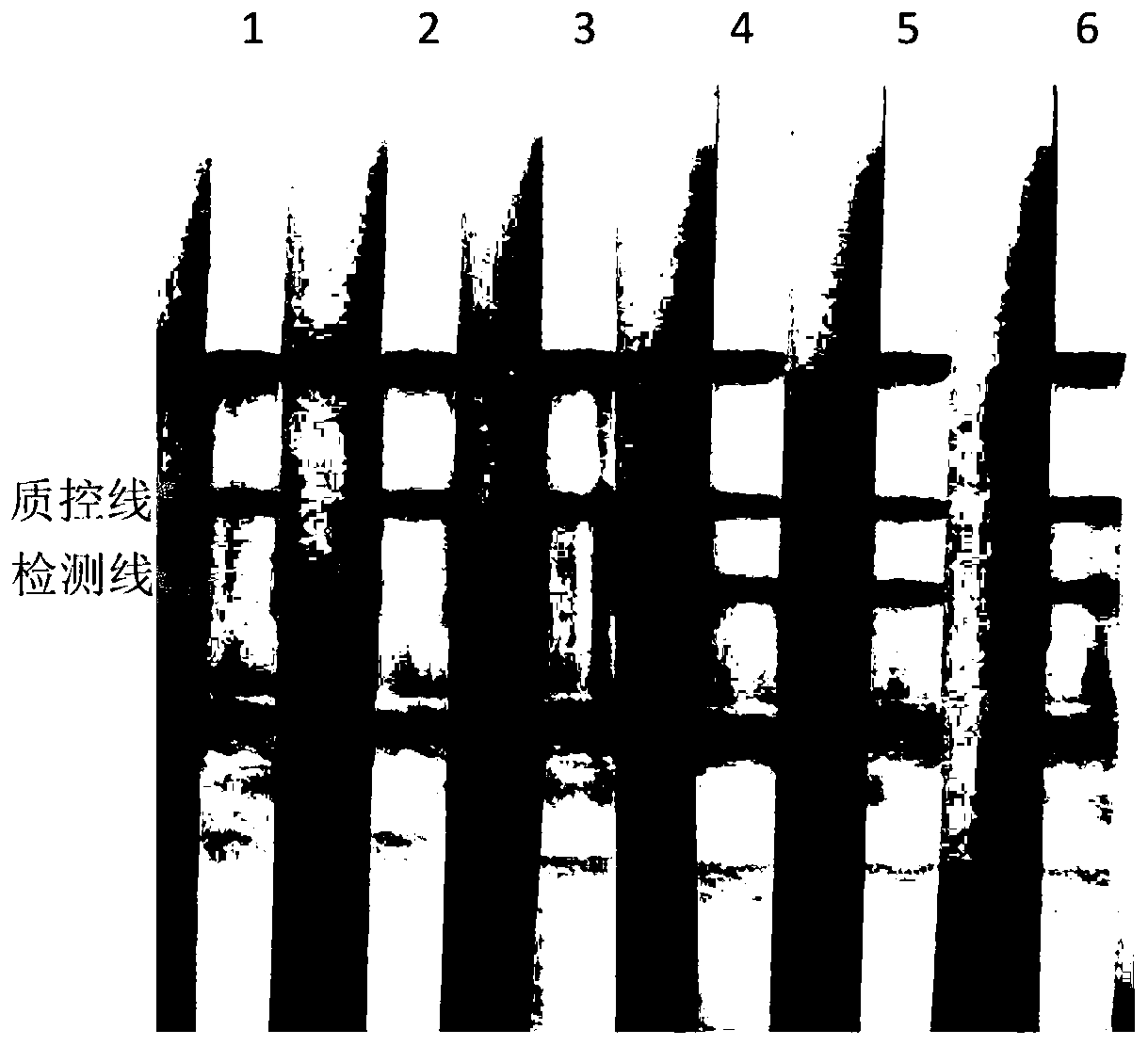
[0050] The Lintan strain of Babesia moiii stored at -20°C was positive, the Babesia moiii strain was positive, the Ningxian strain of Babesia molovi was positive, the Xinjiang strain of Babesia unspecified was positive, and the Babesia unspecified Dunhuang strain was positive. 200 μL of sheep blood that was positive, Theileria reuvei positive, Theileria yoshii positive, and Piroplasma negative was extracted with the QIAamp DNA Mini Kit (Qiagen, Germany), and the specific operation steps were carried out according to the instructions.
[0051] 2. Isothermal Amplification of Cross Primers
[0052] reaction system:
[0053]
[0054] The above reaction tubes were respectively reacted at 63° C. for 60 min.
[0055] 3. Amplified product test strip detection
[0056] Take 5-10 μL of the above reaction product, drop it on the absorbent pad of the chromatograph...
Embodiment 3
[0062] Embodiment 3: the sensitivity test of cross primer amplification
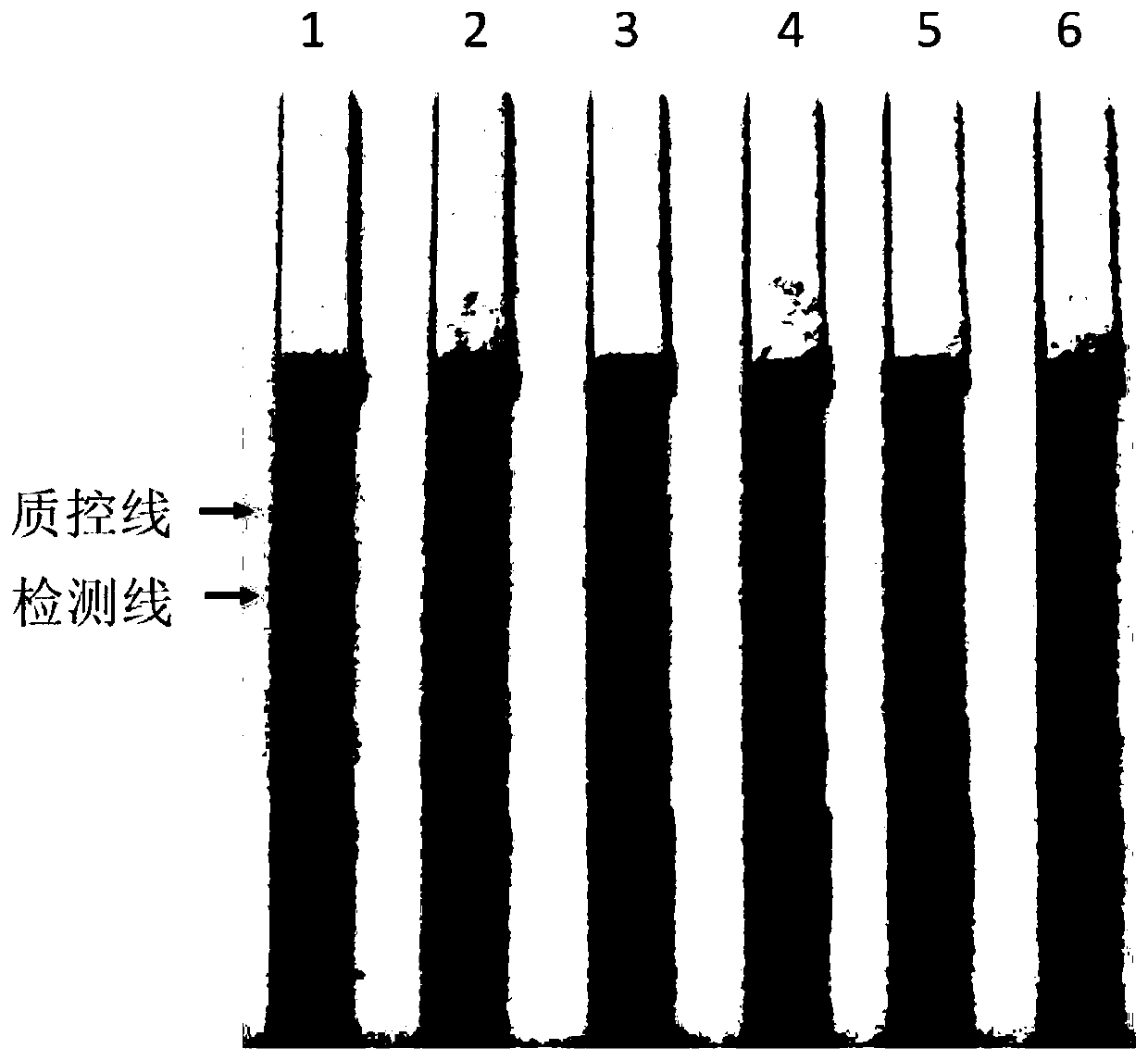
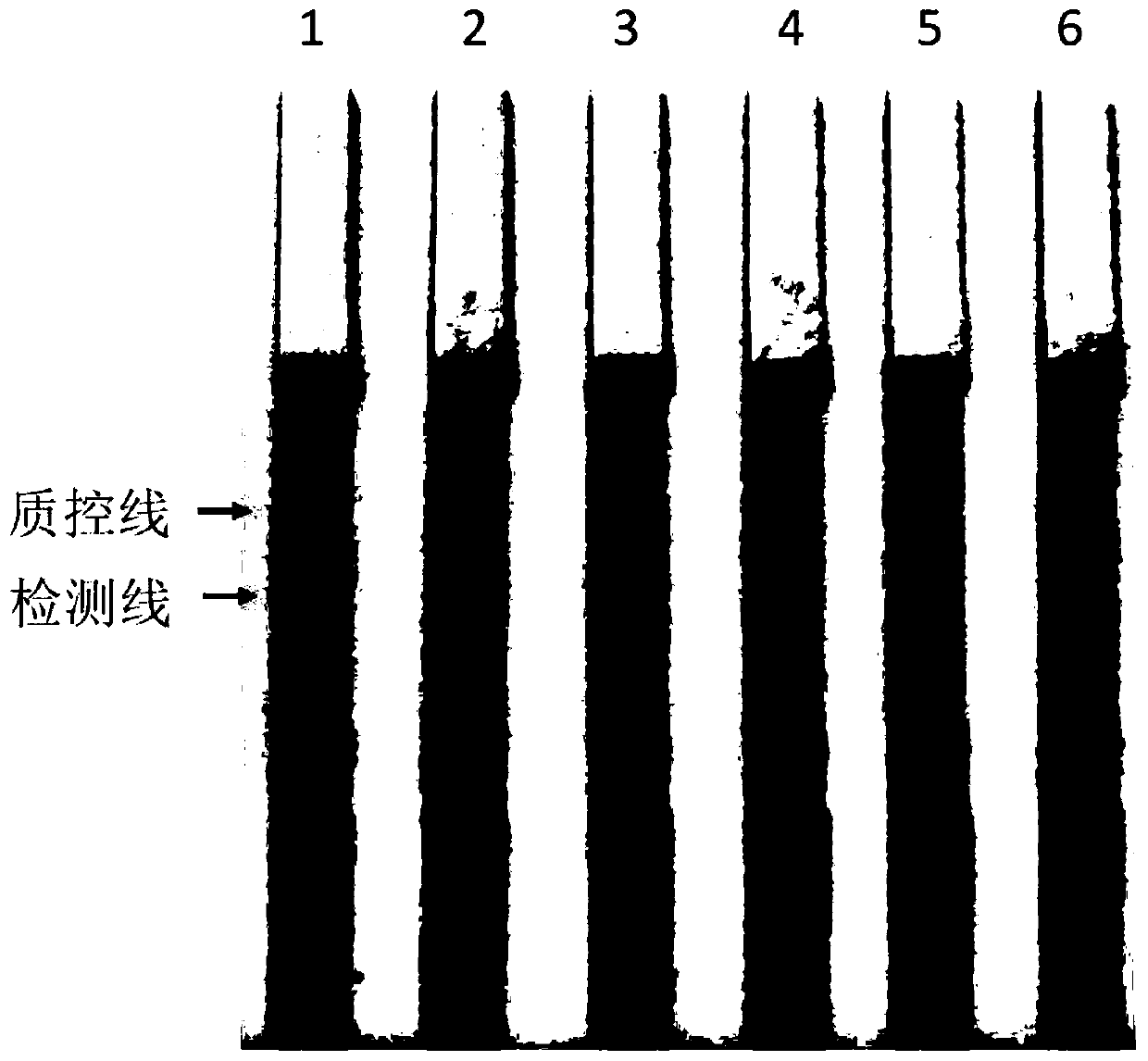
[0063] 1. Babesia unspecified merozoite DNA extraction
[0064] 100 μL of Babesia unspecified merozoites stored at -80°C were extracted with QIAamp DNA Mini Kit (Qiagen, Germany), and the specific operation steps were carried out according to the instructions.
[0065] 2. Preparation of Genomic DNA Gradient Dilution Samples
[0066] The extracted Babesia merozoite genomic DNA samples were determined with a micro-nucleic acid analyzer (NanoDrop2000, Thermo Scientific, USA). The concentration was 50ng / μL, and the genomic DNA samples were serially diluted with ultrapure water to 1ng / μL, 100pg / μL, 10pg / μL, 1pg / μL, and 100fg / μL.
[0067] 3. Isothermal Amplification of Cross Primers
[0068] reaction system:
[0069]
[0070] The above reaction tubes were respectively reacted at 63° C. for 70 min.
[0071] 4. Analysis of amplification products
[0072] Take 5-10 μL of the above reaction product, add i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com