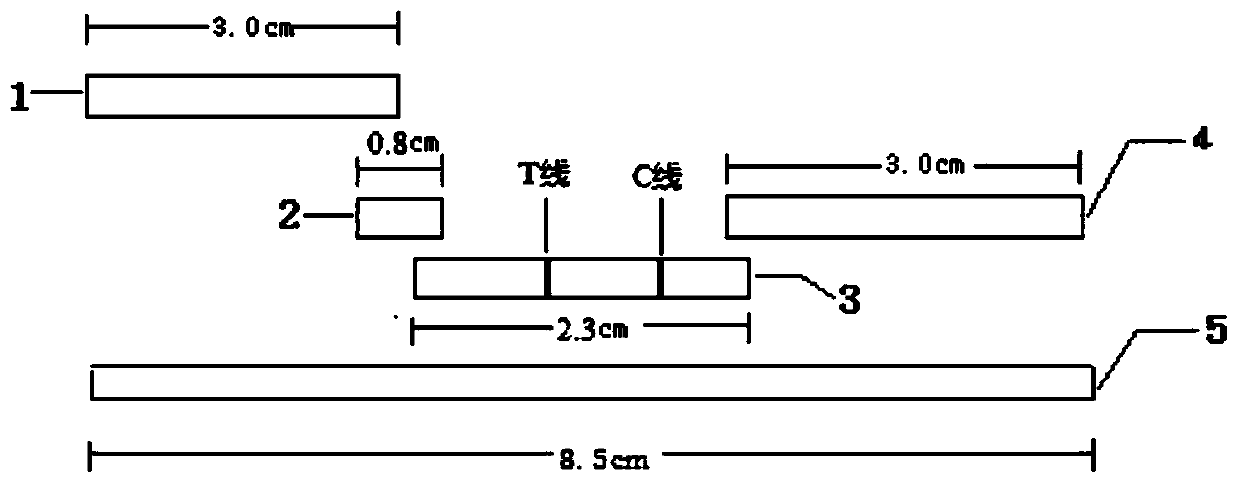
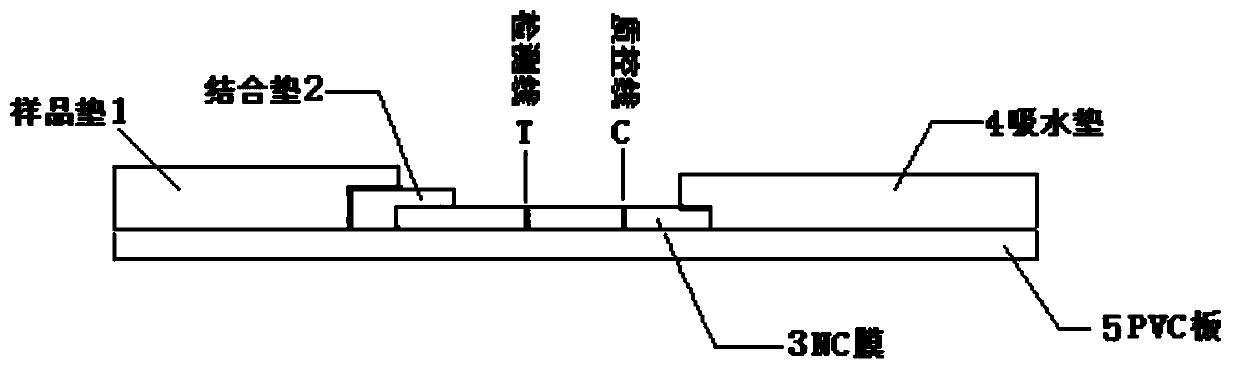
Detection strip for qualitative detection of acinetobacter baumannii specific antibodies in human serum
A technology for qualitative detection of Acinetobacter baumannii, applied in the fields of bioengineering and immunology, can solve the problems of difficult to meet rapid identification, false positives, long time, etc., and achieves low preparation cost, strong antigenicity, and strong capture power. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] The preparation of embodiment 1 Acinetobacter baumannii fusion antigen (Fhue+OmpA+Pilf):
[0056] 1.1) Cloning of Acinetobacter baumannii FhuOmpPil fusion gene
[0057] Obtain the peptides with the most abundant antigenic epitopes in the extracellular domains of the surface proteins Fhue, OmpA and Pilf of Acinetobacter baumannii (the accession numbers in the NCBI protein database are KMV27515, AJF83030 and AJF80497 respectively) and find their gene coding sequences , and optimize its gene coding sequence, and connect the three gene sequences with two coding sequences of flexible connecting peptides (ggsggsggsggs) to form a fusion gene. At the same time, the entire gene sequence was chemically synthesized after introducing the restriction site NdeI at the 5' end of the fusion gene, the termination signal TAA and the restriction site BamHI at the 3' end, which was denoted as FhuOmpPil. The full sequence of the gene and the encoded amino acid sequence are shown in the seq...
Embodiment 2
[0073] Embodiment 2 prepares the latex microsphere marker of mouse anti-human IgG monoclonal antibody
[0074] 2.1) Activation of latex microspheres
[0075] Take 1 mL of a solution of red carboxylated polystyrene latex microspheres (100 nm) with a concentration of 10%, and add 9 mL of 2-(N-morpholino)ethanesulfonic acid (MES) buffer solution (0.1mol / LMES, pH8.5) After mixing; use MES buffer to prepare 10mg / mL N-hydroxysuccinimide (NHS) and 10mg / mL 1-(3-dimethylaminopropyl)-3-ethyl 1-(3- Dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride hydrochloride (EDC) solution;
[0076] Add 1mL of NHS solution and 1mL of EDC solution to the polystyrene latex microsphere (100nm) solution in turn, mix slowly at room temperature for 30 minutes, centrifuge at 19000g for 20 minutes after incubation, remove the supernatant, and use 10mL of borax buffer (0.1 mol / LNa 2 B 4 o 7 , pH8.5) resuspension, oscillation, ultrasonic treatment (ultrasonic breaker product model: YJ92-IIDN, power 50...
Embodiment 3
[0079] The preparation of embodiment 3 binding pads:
[0080] The polyester fiber membrane was cut to a size of 4 cm×0.8 cm / strip, and 64 μL of the latex microsphere marker prepared in Example 2 was added dropwise to the cut membrane. After spraying, dry at 37° C. for 12 hours in an environment with a relative humidity of 20%. Store in airtight and dry place at 25°C.
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle diameter | aaaaa | aaaaa |
| thickness | aaaaa | aaaaa |
| water absorption | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com