Silkworm pupa protein peptide HPP capable of reducing blood fat, and application of silkworm pupa protein peptide HPP
A silkworm chrysalis protein and a blood lipid-lowering technology are applied to the silkworm chrysalis protein peptide HPP and its application fields to achieve the effect of improving the comprehensive utilization level
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
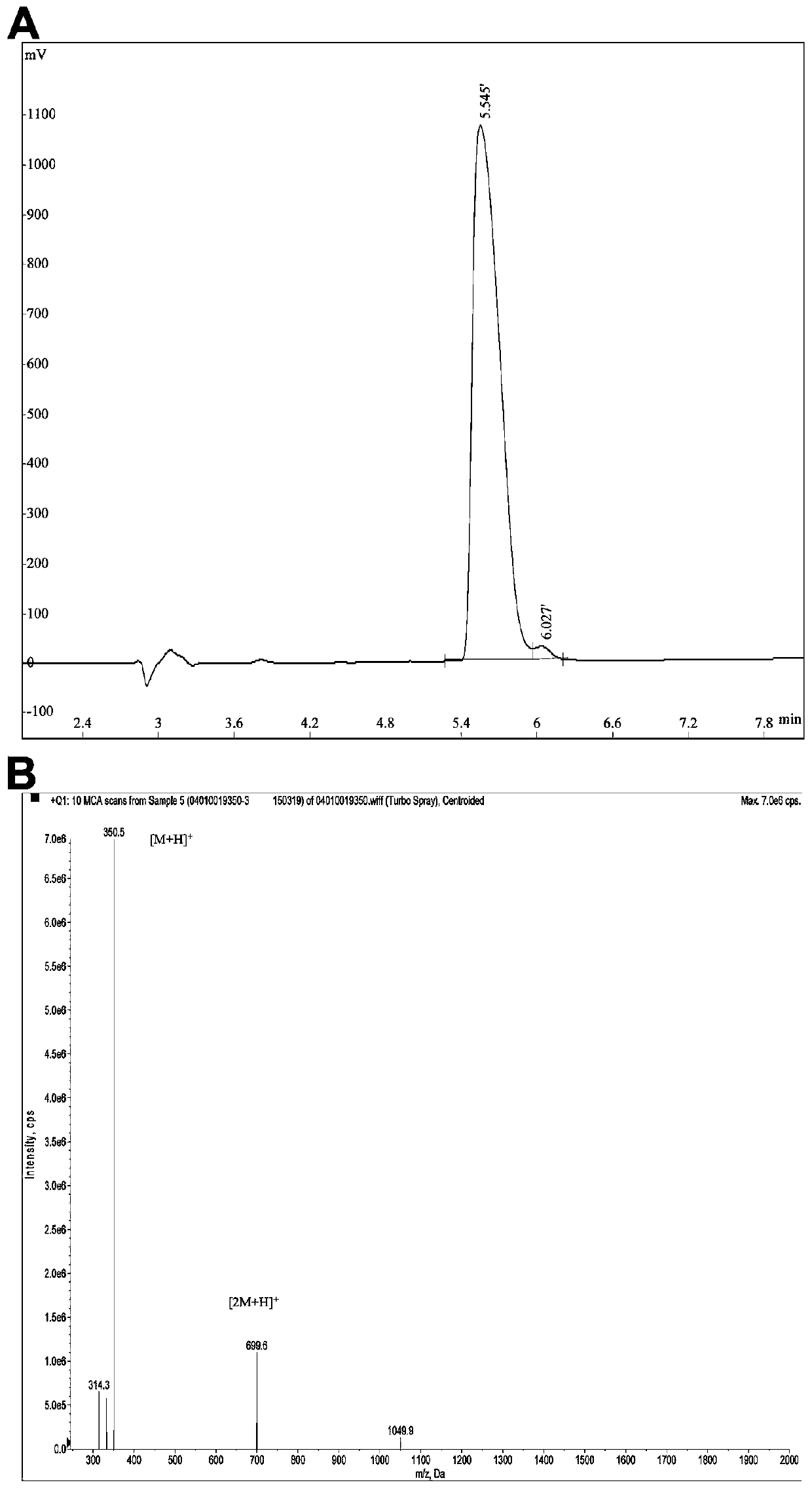
[0027] The acquisition route of silkworm chrysalis protein peptide HPP
[0028] (1) Neutral protease degrades silkworm chrysalis protein
[0029] The enzymatic hydrolysis conditions of silkworm chrysalis protein were studied by using the dual function index of hydrolysis degree and HMGCR inhibition rate. Through response surface optimization experiments, the optimal enzymatic hydrolysis conditions for the preparation of silkworm chrysalis protein lipid-lowering (cholesterol) peptides were obtained: the amount of neutral protease enzyme was 5.1% (w / w), the enzymatic hydrolysis time was 5 hours, the enzymatic hydrolysis temperature was 52°C, and the enzyme The solution pH was 7.0 and the bottom / water ratio (w / v) was 3.9%. The enzymatic hydrolysis solution of silkworm chrysalis protein is obtained.
[0030] (2) Separation and purification of silkworm chrysalis protein enzymatic hydrolyzate
[0031]Peptides with a molecular weight less than 3kDa in the enzymatic hydrolyzate of ...
Embodiment 2
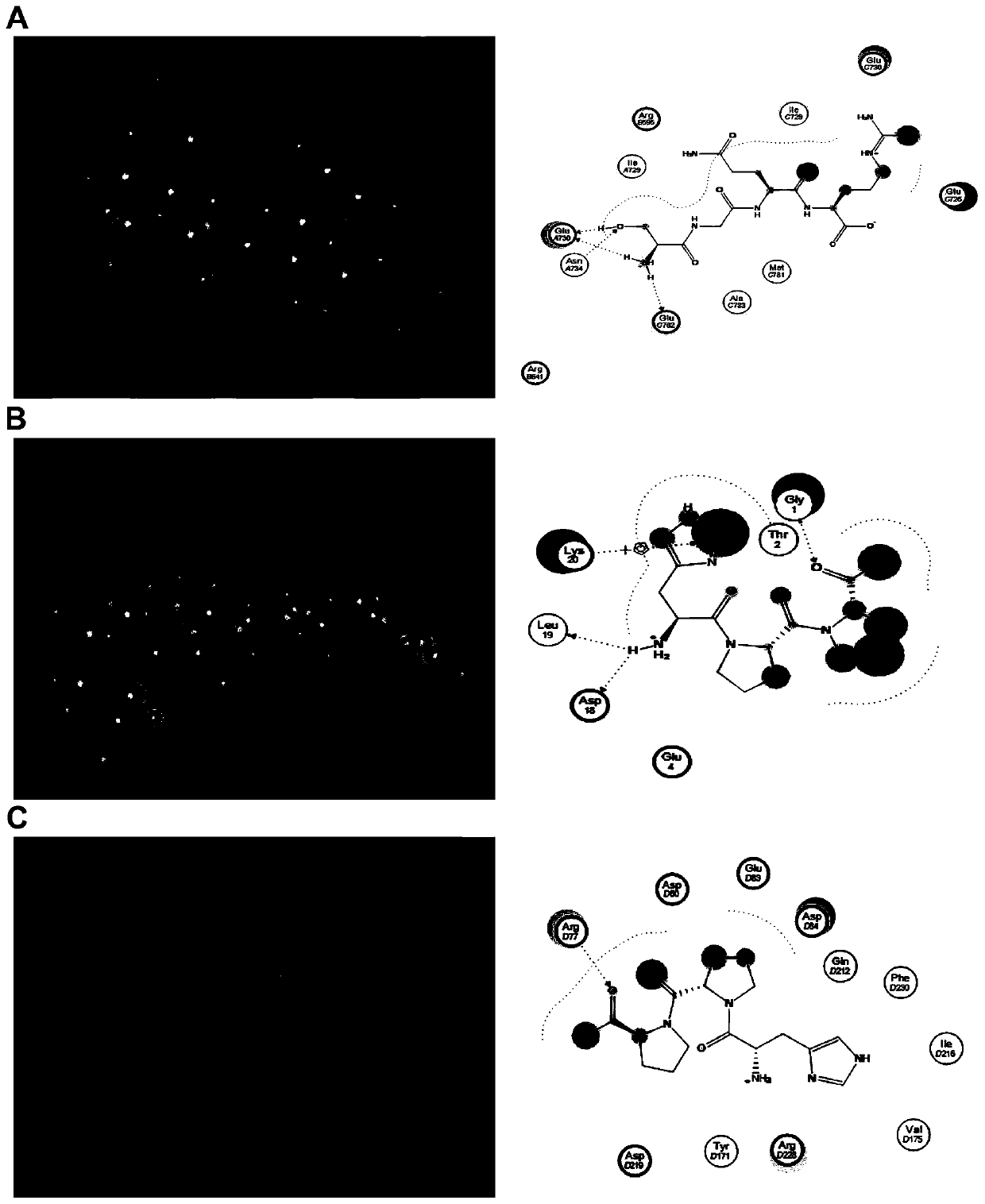
[0049] (1) The molecular docking of HPP and HMGCR was carried out using the Docking module in the MOE software. The HMGCR structure was obtained from the crystal structure in the PDB biomacromolecular structure database (PDB code: 1HW8).
[0050] (2) The molecular docking of HPP and LDLR was carried out by using the Docking module in the MOE software. The HMGCR structure was obtained from the crystal structure in the PDB biomacromolecular structure database (PDB code: 2MG9).
[0051] (3) The molecular docking of HPP and SQS was carried out by using the Docking module in the MOE software. The HMGCR structure was obtained from the crystal structure in the PDB biomacromolecular structure database (PDB code: 3WC9).
[0052] The molecular docking technique was used to analyze the interaction mechanism of HPP with HMGCR, LDLR and SQS. HPP can combine with HMMCR through three hydrogen bonds, which are the three amino acids Arg D595, Arg D641 and Ala A783 in the active pocket of HM...
Embodiment 3
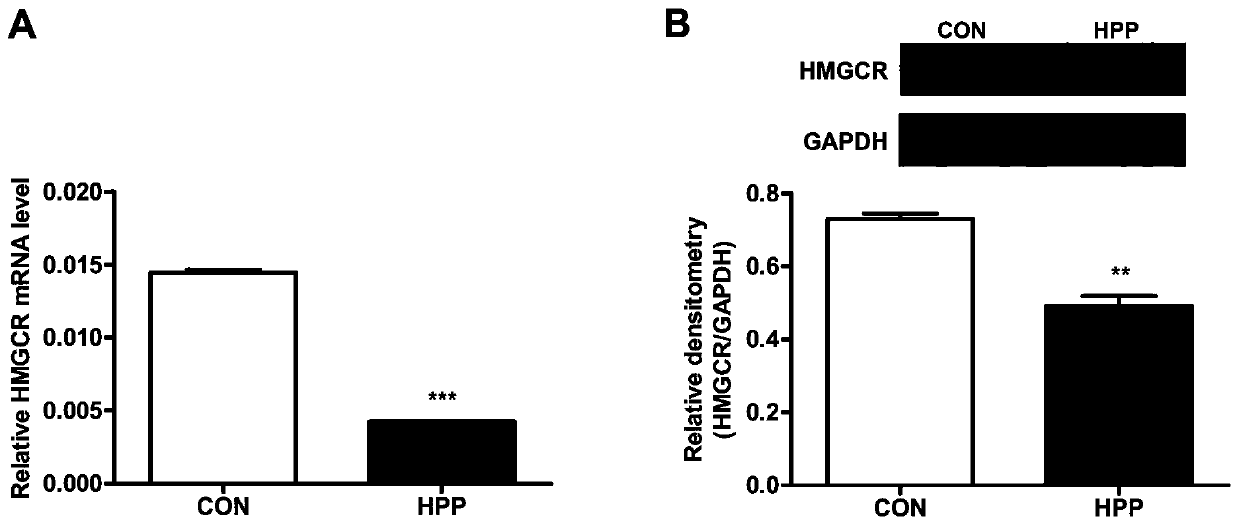
[0054] (1) HepG2 cells were inoculated into 6-well plates, and after 24 hours of culture, they were cultured overnight in the starvation solution. According to different groups, 500ng / mL HPP was not added or added, and the cells were cultured for 8 hours.
[0055] (2) Add 1.0ml Trizol to the cells in each well to lyse the cells, then transfer to a centrifuge tube, add 200μl chloroform; shake vigorously for 15Sec (avoid shaking with an oscillator), let stand at room temperature for 2min, then centrifuge at 12000g for 10min at 4°C; Use a pipette to draw the upper aqueous phase into another clean centrifuge tube, add 600 μL of isopropanol and mix it upside down, centrifuge at 12,000 g at 4°C for 15 min; discard the aqueous phase, add 1 mL of 75% ethanol to wash the precipitate, centrifuge at 12,000 g at 4°C for 5 min, discard The aqueous phase was dried at room temperature for 10 minutes, and the RNA was dissolved in 50 μl of DEPC water, and stored in a -80°C refrigerator for late...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com