Engineering bacterium capable of efficiently expressing chitinase and plant growth promoting application
A high-efficiency expression technology of chitinase, applied in plant growth regulators, plant growth regulators, applications, etc., can solve the problem of low expression and activity of chitinase, and achieve the effect of promoting germination and growth
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Embodiment 1, construction of expression vector and protein expression
[0045] 1. Gene sequence amplification
[0046] A primer pair (ChiA-F, ChiA-R) was designed according to the nucleotide sequence shown in SEQ ID NO.1.
[0047] Table 1: Primers for amplification of chitinase-encoding gene
[0048]
[0049] See Table 1, the underlined part of the forward primer is the Cpo I restriction site, the underlined part of the reverse primer is the Not I restriction site, and the sequence design of this site conforms to T 4 Cohesive ends produced by DNA polymerase trimming.
[0050] PCR reaction system:
[0051]
[0052] PCR reaction conditions: pre-denaturation at 95°C for 5min, denaturation at 95°C for 30s, annealing at 56°C for 30s, extension at 72°C for 1min and 50s, 30 cycles of amplification, and finally extension at 72°C for 10min.
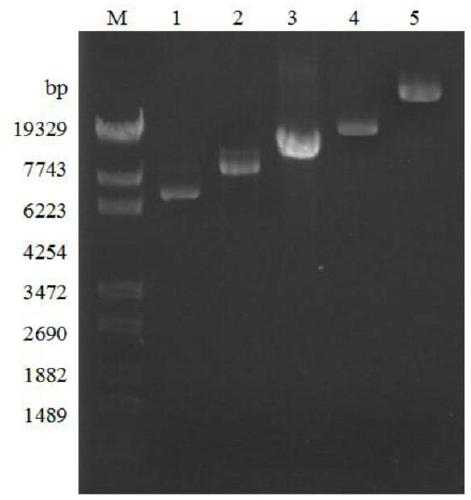
[0053] Detect the PCR product with 0.7% agarose gel electrophoresis, and use DNA purification kit (Omega company)
[0054] 2....
Embodiment 2
[0065] Example 2, construction of cofactor genes and related vectors
[0066] 1. Cofactor gene amplification
[0067] According to the HAC1, ERV29, SEC16, COG5 and TRM1 cofactor gene sequences derived from Pichia pastoris GS115, primers were designed for gene amplification, and these genes were respectively constructed between the Xho I and Xba I restriction sites of the pGAPZB vector.
[0068] See Table 2, the underlined part of the forward primer is the Xho I restriction site and its homologous sequence at the left end of the vector, and the underlined part of the reverse primer is the Xba I restriction site and its homologous sequence at the right end of the vector , the sequence design of this site conforms to T 5 A method for constructing vectors with exonucleases.
[0069] Table 2: Primers for Cofactor Gene Amplification
[0070]
[0071] PCR reaction system:
[0072]
[0073] PCR reaction conditions: 98°C pre-denaturation for 5 minutes, 98°C denaturation for 1...
Embodiment 3
[0078] Example 3, cofactor and 4 copies of chitinase co-expression recombinant strain
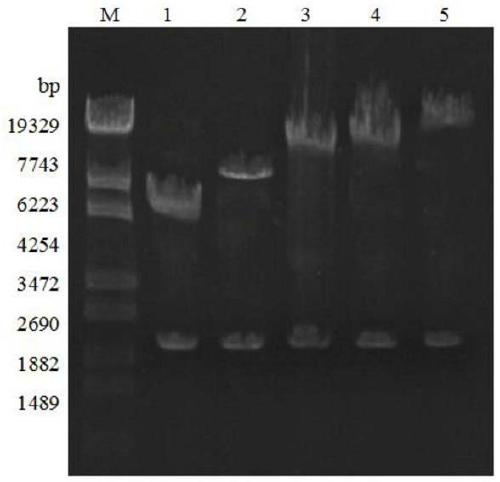
[0079] After the plasmids pGAPZB-HAC1, pGAPZB-ERV29, pGAPZB-SEC16, pGAPZB-COG5, and pGAPZB-TRM1 were linearized and digested with Avr II, the digested products were recovered from the solution. Then, under the condition of 1.5kv, electrotransform the GS115 competent with 4 copies of chitinase gene, spread the YPD plate containing 100 μg / mL Zeocin, and incubate at 28°C for 2 days. Then transfer the bacteria on the YPD (Zeocin) plate to the YPD plate and mark it. A yeast colony PCR was then performed to narrow the screen. An appropriate amount of the screened bacteria was used for methanol-induced expression in shake flasks. Subsequent expression steps of chitinase were carried out in accordance with point 5 in Example 2 for methanol-induced expression.
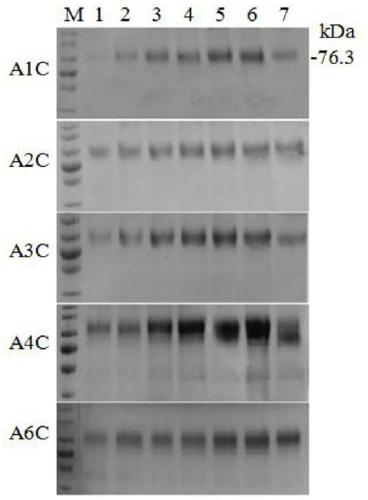
[0080] The result is as Figure 4 Shown: 1 is the control with 4 copies of chitinase, 2 / 3 / 4 / 5 / 6 are the chitin when overexpressing HAC1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com