Method for preparing serratia nuclease through efficient secretory fusion expression and recombination in methylotrophic yeast
A fusion expression, methanol yeast technology, applied in the field of bioengineering, can solve the problems such as the inability to effectively block the biological activity of the target protein, the lack of an extracellular secretion system in Escherichia coli, the high toxicity of host cells, etc. high-efficiency expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1. Preparation of prodigal nuclease based on HV1 fusion protein
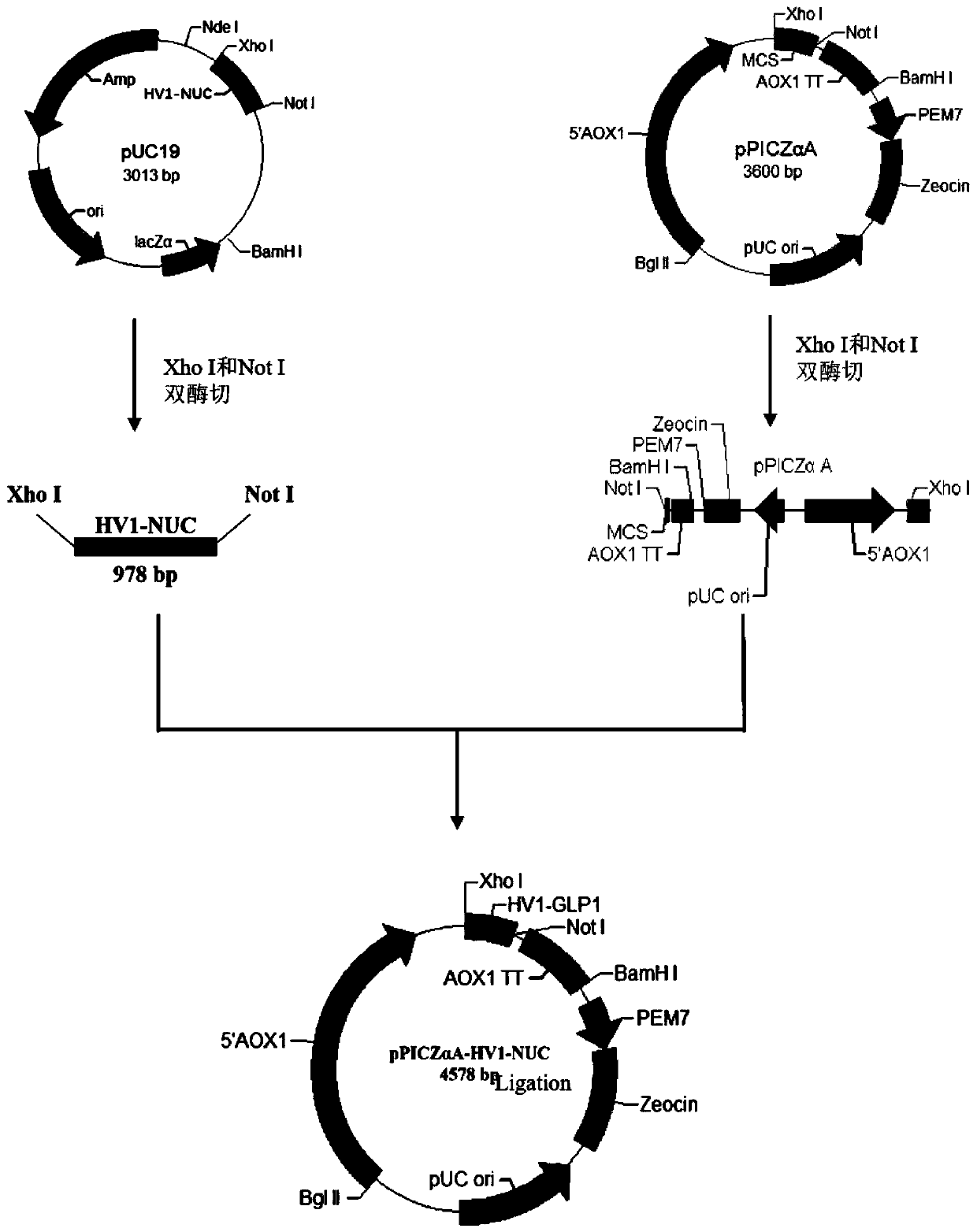
[0037] (1) Construction of fusion vector pPICZαA-HV1-NUC
[0038] ①Fusion protein sequence design
[0039] In this example, the N-terminus of the fusion protein involved is the HV1 sequence (SEQ ID No: 1), the middle is the enterokinase cleavage site sequence (DDDDK), and the C-terminus is the prodigal nuclease (SEQ ID No: 4 ). The complete sequence of the fusion protein (SEQ ID No: 5) is:
[0040] VVYTDCTESGQNLCLCEGSNVCGQGNKCILGSDGEKNQCVTGEGTPKPQSHNDGDFEEIP
[0041]
[0042] The underlined part is the amino acid sequence of the HV1 mutant, the double underlined part is the amino acid sequence of the enterokinase cleavage site, and the underlined part is the NUC amino acid sequence.
[0043] ②Fusion gene design and synthesis
[0044] According to the above amino acid sequence, the nucleotide sequence was designed according to the codon preference of Pichia pastoris. Design a stop codon ...
Embodiment 2
[0057] Example 2. Preparation of prodigal nuclease based on HV2 fusion protein
[0058] (1) Construction of fusion vector pPIC9K-HV2-NUC
[0059] ①Fusion protein sequence design
[0060] In this example, the N-terminus of the fusion protein involved is the HV2 sequence (SEQ ID No: 2), the middle is the enterokinase cleavage site sequence (DDDDK), and the C-terminus is the prodigal nuclease (SEQ ID No: 4 ). The complete sequence of the fusion protein (SEQ ID No: 7) is:
[0061] VVYTDCTESGQNLCLCEGSNVCGQGNKCILGSDGEKNQCVTGEGTPKPQSHNDGDFEEIP
[0062]
[0063] The underlined part is the amino acid sequence of the HV2 mutant, the double underlined part is the amino acid sequence of the enterokinase cleavage site, and the underlined part is the NUC amino acid sequence.
[0064] ②Fusion gene design and synthesis
[0065] According to the above amino acid sequence, the nucleotide sequence was designed according to the codon preference of Pichia pastoris. Design a stop codon s...
Embodiment 3
[0078] Example 3. Preparation of non-hirudin fusion protein by prodigal nuclease
[0079] (1) Construction of vector pPICZαA-NUC
[0080] ①Protein sequence design
[0081] In this embodiment, the amino acid sequence of Prodigobacterium nuclease NUC (SEQ ID No: 4) is:
[0082]
[0083] ② Gene design and synthesis
[0084] According to the above amino acid sequence, the nucleotide sequence was designed according to the codon preference of Pichia pastoris. Design a stop codon sequence behind the expression sequence, and design XhoI and NotI restriction enzyme sites at the N-terminal and C-terminal of the sequence, respectively, and entrust the whole gene synthesis.
[0085] ③The NUC sequence synthesized by the whole gene was double-digested with XhoI and NotI and then inserted into the pPICZαA vector that had been digested with the same enzyme and transformed into Escherichia coli DH5α or Top10 cloning host bacteria. Positive clones were picked, and the recombinant plasmid ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com